Ugly Little Bits of the Data Science Process
Eduardo Ariño de la Rubia2016-06-09 | 11 min read
This morning there was a great conversation on Twitter, kicked off by Hadley Wickham, about one of the ugly little bits of the data science process.
During a data analysis, you'll often create lots of models. How do you name them? Who has written good advice on the subject?
— Hadley Wickham (@hadleywickham) June 8, 2016
I found the question incredibly interesting. It’s one of those weird dark corners where data science and the reality of computers and file systems bump up. Almost anyone who has ever done data science has found themselves with a folder looking something like this:
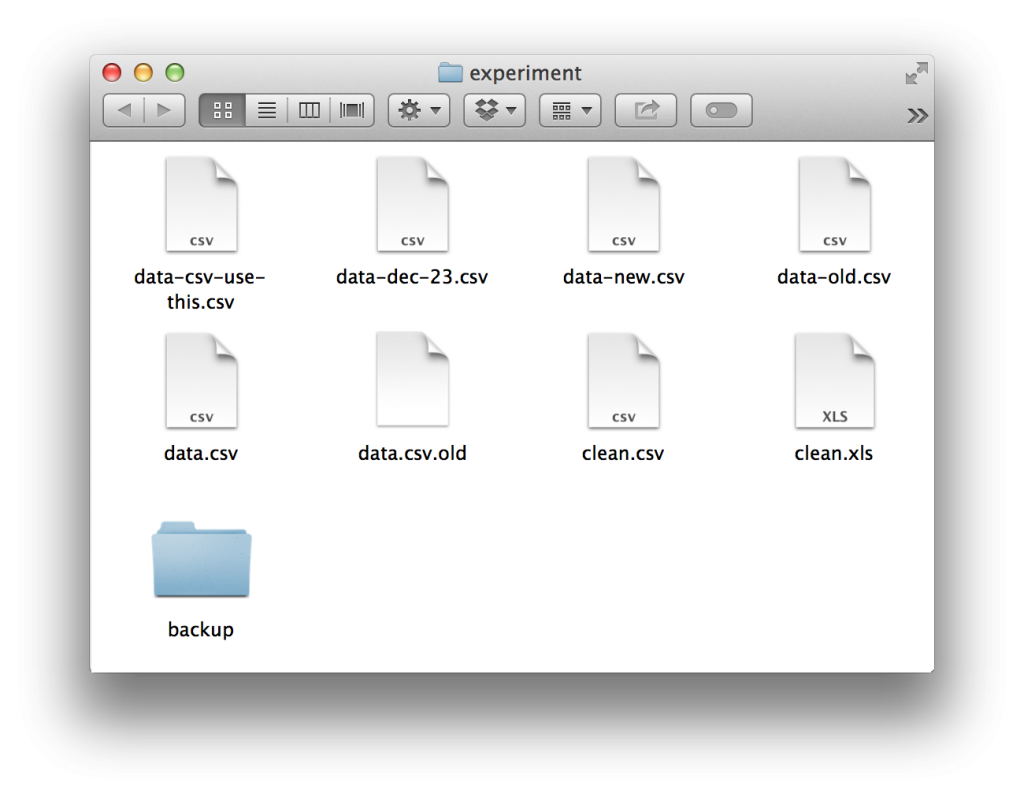
Everyone starts out with the best of intentions regarding how they want their project to be organized. Yet without a systematized method of keeping revisions and experiments, entropy will almost always guide your hand towards chaos. There were a number of responses to Hadley on twitter, almost all of them with some sort of variation on the theme: “store some metadata in the file name.”
They ranged from the relatively straightforward, such as Oliver Key’s (@quominus) idea that models should be stored as [model type] + [name of the dataset]
The complexity escalated a bit, as H2O’s ML and ensembling expert Erin Ledell (@ledell) suggested something along the lines of [lib/algo]+[dataset]+{some,key,params}+[md5hash of model object]. However, she acknowledged this only provided a limited snapshot as she added: “Too long to put all params in the filename.”
Something like [lib/algo]+[dataset]+{some,key,params}+[md5hash of model object]. Too long to put all params in the filename.
— Erin LeDell (@ledell) June 8, 2016
To the ingenious but unfortunately completely untenable suggestion of UBC professor (and best twitter follow of 2016) Jenny Bryan (@JennyBryan) that it would be nice if the system could store “some suitably transformed version of the actual formula.” Which she acknowledged was probably “super ugly.”
it would be super ugly but how about some suitably transformed version of the actual formula? 1/2
— Jenny Bryan (@JennyBryan) June 8, 2016
These are some of the sharpest, smartest people that I know, all answering a pretty tough question. I think they would probably all agree that the answers are OK, but leave something to be desired. When that much brainpower is unable to come up with a satisfactory solution, it’s probably a good time to ask if the question being asked is the right question.
Rationalizing the Question
During a data analysis, you'll often create lots of models. How do you name them? Who has written good advice on the subject?
Hadley’s original question packs quite a bit of complexity into less than 140 characters. It acknowledges the constant iterative nature of the exploratory and analysis phase of the data science process. In this phase, the data scientist often doesn’t know the right question to ask, nor what the limitations of her data set are. This inspires a cycle of rapid exploration and hypothesis generation, in which many different lenses are applied to the data. Most of these are going to be exploratory dead ends, but the DS may not know this during the moment. This leads to the desire to “save them”, just in case. This is the second part of Hadley’s tweet “How do you name them.”
I think it’s interesting to note how tools shape the way we think about problems around us. In the mental model that presupposes this question, predictive models are an outcome of a training experiment, to be serialized as artifacts into a file system. This mental model prunes a lot of possible ways to look at this problem, including the way Domino handles this scenario. When I read Hadley’s tweet, I realized this was a problem I had not faced in the last 3 years, since I became a dedicated Domino user, and felt a quick exploration was worthwhile.
An Alternate Approach
I would challenge that "saving a model" is unlikely to be what a data scientist really wants in this process. Data Scientists want to be able to generate as many different hypotheses as they want, and be able to reproduce and select any of them moving forward in their process. A model, by itself, is an artifact that was created with the coming together of a number of components. At Domino, one of our core pieces of technology is our reproducibility engine. This key technology drives our ability to become the “system of record” for model experimentation and creation in an organization. It captures everything about an experiment and saves it so that it can be later studied, analyzed, compared, or reproduced. We define an experiment as follows:
experiment = dataset + code + hyperparameters + compute environment + results
Everything that is run inside of the Domino environment is run inside of our reproducibility engine, and for every run, we capture this immutable tuple. This frees the data scientist from worrying about how to save her model, what name to give it, or when she needs it 6 months down the road, remembering how it is she generated it in the first place.
An Example
All of this is pretty abstract, so I created a simple example that should make this all pretty clear. I have created a sample project on Domino called "evaluate_model".
This project contains a single file, evaluate_model.R. The purpose of this script is to use Max Kuhn’s amazing caret package to train a model, on a data set, with a given algorithm, and generate some diagnostic criteria. This file when run takes 3 parameters:
- Model Type: This is any of the caret model types (rf, nnet, nb, glmnet, etc…)
- Data Set: This is any of R’s built-in data sets (
iris,mtcars, etc…) - Target: This is a categorical column in the above data set which the model will attempt to predict.
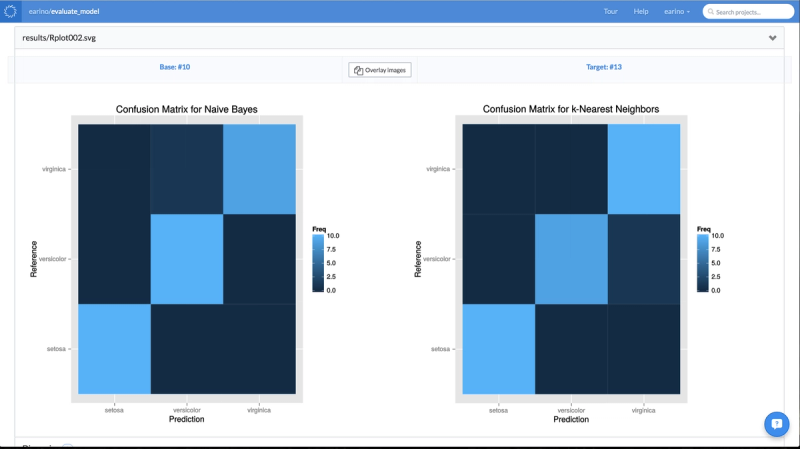
It then trains the model, on that dataset, to predict the desired target column. It generates some basic diagnostics, as well as some nice diagnostic plots. In particular, it generates a confusion matrix visualization using ggplot.
Let’s say we want to take the venerable iris dataset, and predict the Species column from all the other predictors. We want to do this for the following model types: random forest, neural network, glmnet, and knn. If we are doing this without Domino, the best we can do is save model files like for each experiment:
glmnet-iris.RData, nnet-iris.RData, if we follow Oliver’s suggestion, etc… if we follow Erin’s suggestion- I honestly don’t actually know what it would look like for Jenny’s suggestion
This pollutes the project with a bunch of model files, almost all of which we will end up discarding. However, with Domino, we can execute all of these experiments in parallel across multiple machines (train a random forest, neural network, glmnet, and knn), save all of the models as model.RData, and the reproducibility engine captures the entire experiment in a fully reproducible fashion that we can look up by diagnostic statistic. What was previously a folder with a bunch of files, now becomes this:
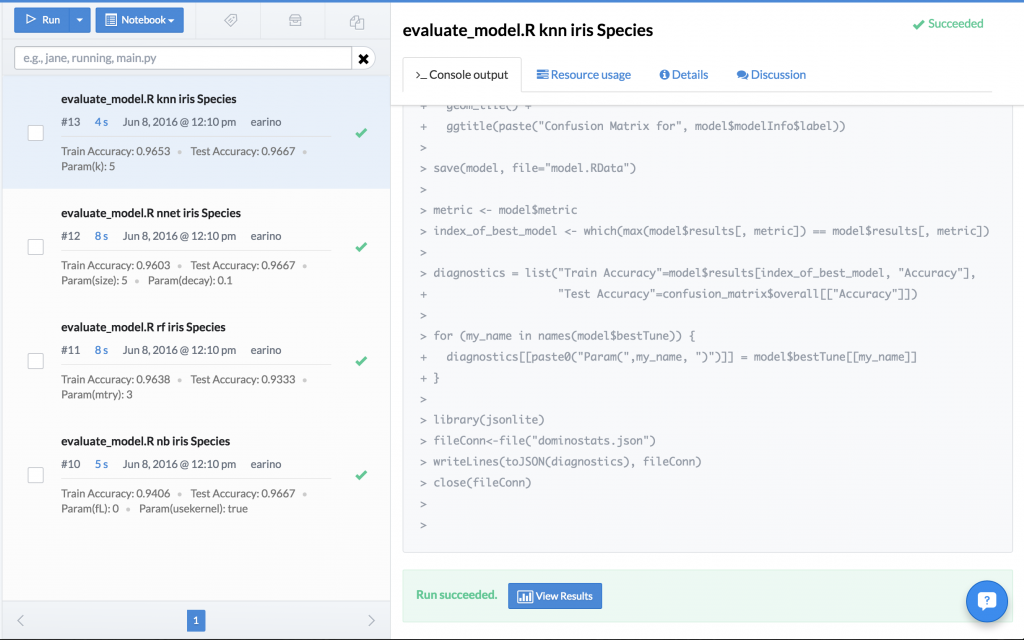
All of the models, the tuning parameters that were used in the hyperparameter search, and the results are all stored inside of the reproducibility engine and visible in the column on the left. The data scientist can now pick which model fits her needs best, and simply use that experiment as the base for her future work. No worrying about files, overwriting old results, anything. It’s all handled by the Domino environment. However, there’s an even more powerful feature available. You may remember that the script generated some diagnostic plots. Domino allows you to do a side-by-side comparison of these experiments to further inform your experimental decisions. Domino allows us to compare the confusion matrix visualization or any of the results generated by your experiment:
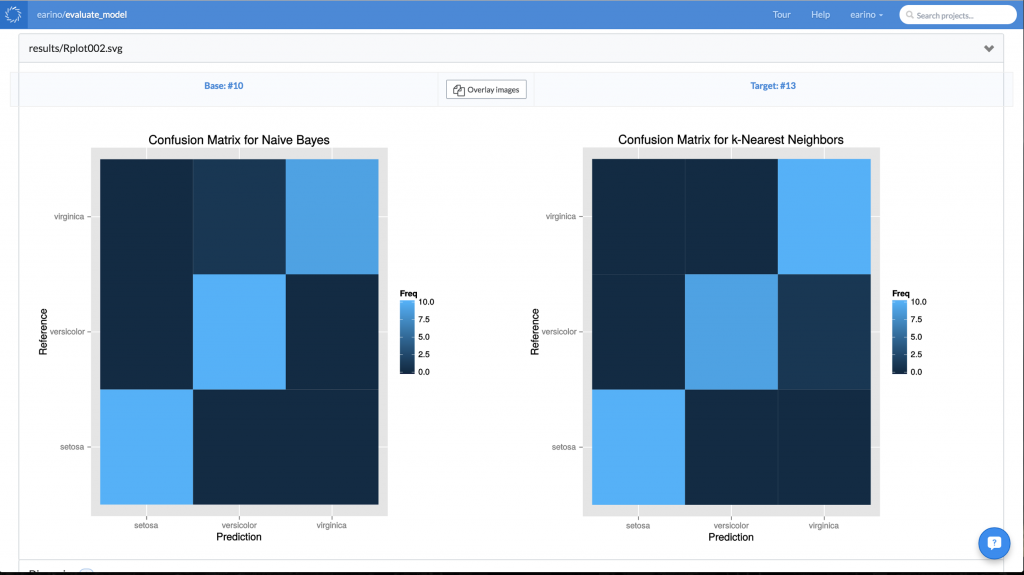
Or even information about how each model was tuned during the hyperparameter search:
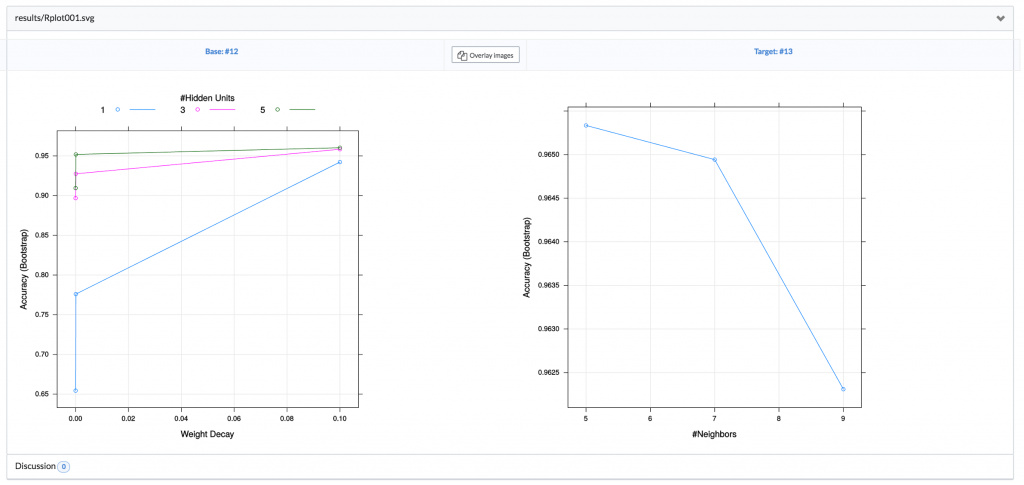
This is not something that would ever be possible when saving files to a project folder on your filesystem. We cover a lot of this functionality in our post Unit Testing for Data Science.
Conclusion
This post started out talking about how to name files, and the difficulties in how you pack all of the information about an experiment that generated a model, into a filename. That’s a tough problem that’s probably unsolvable, as the number of variations, hyperparameters, and assumptions you would want to bake into a filename would quickly become unwieldy. Since I started using Domino, I realized that what I wanted wasn’t good filenames, it was fully reproducible, indexed, searchable, discoverable records of my experiments. Records that made it easy to compare the behavior of models and algorithms against each other fluidly, and make it easy for me to make a decision about which approach to take forward through the rest of the data science process. Using Domino’s reproducibility engine, and the automated experiment tracking capabilities, I can fire up as many different variations of an experiment I want, be confident that all of the information about the experiment will be saved, make a decision about which path to take forward, and with a single mouse click use it as the foundation for future work.
I will be giving a talk at useR 2016 on a related topic titled Providing Digital Provenance: From Modeling to Production. In this talk, I will present a solution we have developed at Domino that allows for every model in production to have full reproducibility: From EDA to the training run, and the exact datasets which were used to generate the model. We discuss how we leverage Docker as our reproducibility engine, and how this allows us to provide the irrefutable provenance of a model. Make sure to come by and say hello if this is interesting to you!
Eduardo Ariño de la Rubia is a lifelong technologist with a passion for data science who thrives on effectively communicating data-driven insights throughout an organization. A student of negotiation, conflict resolution, and peace building, Ed is focused on building tools that help humans work with humans to create insights for humans.
Subscribe to the Domino Newsletter
Receive data science tips and tutorials from leading Data Science leaders, right to your inbox.
By submitting this form you agree to receive communications from Domino related to products and services in accordance with Domino's privacy policy and may opt-out at anytime.



