Using PySpark and PCA to analyze large neuroimaging datasets
Sergul Aydore, Ph.D., and Syed Ashrafulla, Ph.D.2021-08-19 | 11 min read

Principal component analysis in neuroimaging data using PySpark
In this post we will describe how we used PySpark, through Domino's data science platform, to analyze dominant components in high-dimensional neuroimaging data. We will demonstrate how to perform Principal Components Analysis (PCA) on a dataset large enough that standard single-computer techniques will not work.
PCA is a widely used tool in machine learning, data analysis, and statistics. The goal of PCA is to represent the data in a lower dimensional space. PCA is useful for denoising, as well as for data exploration of dominant linear trends in the data. As part of the setup, PCA ensures that the dimensions returned are orthonormal and capture the largest directions of variance in the data.[2]
What is principal component analysis (PCA)?
To describe PCA, we will start with a data set that has m features and n observations, represented as a matrix X with m rows and n columns. Let's also assume we've subtracted a constant from each feature so that the resulting features all have mean 0.
Our goal is to find the d vectors that best capture the variation in the dataset across observations, represented as a matrix Z with d uncorrelated rows and n columns. There are two ways to find this summarization of the data: Using the sample covariance matrix, and using Singular Value Decomposition (SVD).
PCA using the covariance matrix
Specifically, we will use the eigenvalue decomposition of the sample covariance matrix. The sample covariance matrix is \begin{equation}C_{xx} = \frac{1}{n}X X^T\end{equation} where $C_{ij}$ entry is the covariance between $i$-th feature with $j$-th feature. Our goal is to find a matrix $P$ that projects $X$ onto a lower-dimensional subspace
\begin{equation}
Z = \mathbf{P}X \label{eqn:projection}
\end{equation}
where the covariance matrix of $Z$, $C_{zz}$, is diagonal (equivalently, the rows of $\bZ$ are orthogonal to each other):
\begin{equation}
C_{zz} = \frac{1}{n}Z Z^T = P C_{xx} P^T = D \label{eqn: diagonal_condition}
and $$D_{ij} = 0$ $for $$i \neq j$$.\end{equation}
Since $$C_{xx}$$ is a symmetric matrix the eigenvalue decomposition can be represented by:
\begin{equation}
C_{xx} = E D E^T
\end{equation}
where E is an orthogonal matrix, each column an eigenvector of $$C_{xx}$$. Plugging into {eqn: diagonal_condition}, we can see that setting $$P = E^T$$ satisfies the conditions on D. Hence we get our principal components by setting $$Z = E^T X$$.
PCA using singular value decomposition
We don't need to use the sample covariance matrix to get the largest principal components. Define $$Y = \frac{1}{\sqrt{n}}X^T$$; as a result, each column of $\bY$ has zero mean. From linear algebra we can decompose $\bY$ into three matrices
\begin{equation}
Y = U S V^T
\end{equation}
where $\bU$ is $n \times k$, and $\bV$ is $k \times $$m$$ and $$S$$ is $k \times k$, for some $k$. $k$ is called the rank of the matrix. If we carefully look at the product $$Y^TY$$
\begin{align}
Y^TY &= \frac{1}{n} X X^T = C_{xx} \
&= V S^2 V^T
\end{align}
the columns of V are not only the eigenvectors of $Y^TY$; they are also the eigenvectors of $C_{xx}$. Therefore, V can be the projection matrix used to compute $Z$: $Z = V^T X$.
Singular value decomposition is often preferred over eigendecomposition of the covariance matrix because the calculation of the covariance matrix is a source of error. In singular value decomposition, with such a large dataset, we are much more robust to errors due to dynamic range of numbers or computational error.
Toy example
Covariance matrix
Let's start with a toy example
\begin{equation}
X = \begin{array}{ccc} 0 & 2 & 4 \ 1 & 0 & 0 \ 0 & 3 & 0 \ 7 & 4 & 6 \ 0 & 5 & 7 \end{array}
\end{equation}
where each column is another observation of five dimensional random variable. Let's compute the sample covariance matrix of $\bX$:
\begin{align}
C_{xx} = \frac{1}{3} \left( X - {\mu} \right) \left( X - {\mu} \right)^T
\end{align}
where five-dimensional vector
\begin{align}
{\mu} \mbox{\stackrel{\triangle}{=}} \left[ \begin{array}{ccccc} 2 & 0.33 & 1 & 5.6 & 4 \end{array} \right]^T
\end{align}
is the mean across all observations. Hence the covariance matrix is:
\begin{align}
C_{xx} = \left[ \begin{array}{ccccc} 2.66 & -0.66 & 0 & -0.66 & 4.66 \
-0.66 & 0.22 & -0.33 & 0.44 & -1.33 \
0 & -0.33 & 2 & -1.66 & 1 \
-0.66 & 0.44 & -1.66 & 1.55 & -2 \
4.66 & -1.33 & 1 & -2 & 8.66 \end{array} \right]
\end{align}
and eigen value decomposition is:
\begin{equation}
C_{xx} = E D E^T
\end{equation}
where
\begin{align}
E = \left[ \begin{array}{cc} -0.44 & -0.28 \
0.13 & -0.05 \
-0.12 & 0.76 \
0.21 & -0.56 \
-0.84 & -0.11 \end{array} \right]
\end{align}
and
\begin{align}
D = \left[ \begin{array}{cc} 12 & 0 \ 0 & 3.1 \end{array} \right].
\end{align}
There are 2 eigenvectors because the rank of $X$ is 2 and each column of $\bE$ represents an eigenvector which are ordered by the magnitude of eigen values represented in diagonal elements of matrix $\bD$. If we want to represent $\bX$ in 2-dimensional space, all we need to do is to multiply $X$ by $E^T$:
\begin{align}
Z &= P X \label{eqn:Z} \
&= E^T X \
&= \left[ \begin{array}{ccc} 1.64 & -4.64 & -6.42 \
-4.01 & -1.11 & -5.33 \end{array} \right].
\end{align}
If you want your output to be zero mean across columns, you can multiply $E^T$ by $X - m$:
\begin{align}
Z_{{zeromean}} &= E^T \left(X - m \right) \label{eqn:Zzeromean} \
&= \left[ \begin{array}{ccc} 4.79 & -1.5 & -3.28 \
-0.52 & 2.37 & -1.84 \end{array} \right].
%&= \left[ \begin{array}{ccc} 4.79 & -1.5 & -3.28 \
% -0.52 & 2.37 & -1.84 \end{array} \right].
\end{align}
Singular value decomposition (SVD)
Following our notation in section 1.2, the matrix $Y$ is:
\begin{align}
Y &= \frac{1}{\sqrt{t}} X^T \
&=\frac{1}{\sqrt{3}} \left[ \begin{array}{ccccc} 0 & 1 & 0 & 7 & 0 \
2 & 0 & 3 & 4 & 5 \
4 & 0 & 0 & 6 & 7 \end{array} \right].
\end{align}
Computing SVD results:
\begin{equation}
Y = U S V^T
\end{equation}
where $U = \left[ \begin{array}{cc} 0.79 & -0.17 \ -0.25 & 0.77 \ -0.54 & -0.60 \end{array} \right]$, $\bD = \left[ \begin{array}{cc} 3.46 & 0 \ 0 & 1.76 \end{array} \right]$ and $\bV = \left[ \begin{array}{cc} -0.44 & -0.28 \
0.13 & -0.05 \
-0.12 & 0.76 \
0.21 & -0.56 \
-0.84 & -0.11 \end{array} \right]$.
Note that matrices $V$ and $E$ are equivalent, as discussed in section 1.2.
Implementing PCA
Using scikit-learn
The code below demonstrates PCA using scikit-learn. The results are consistent with theoretical results we showed in section 2 except the sign of the components. This is not an issue because sign does not affect the direction of the components.
import numpy as np
from sklearn.decomposition import PCA
x = np.array([[0.0, 1.0, 0.0, 7.0, 0.0], [2.0, 0.0, 3.0, 4.0, 5.0],
4.0, 0.0, 0.0, 6.0, 7.0], dtype=object)
pca = PCA(n_components=2, whiten=False)
pca.fit(x)PCA(copy=True, n_components=2, whiten=False)Components (matrix V)
pca.components_.transpose()array([[-0.44859172, -0.28423808],
[ 0.13301986, -0.05621156],
[-0.12523156, 0.76362648],
[ 0.21650757, -0.56529588],
[-0.84765129, -0.11560341]])Representation in 2-D (matrix Z)
pca.transform(x)array([[ 4.79037684, -0.5239389 ],
[-1.50330032, 2.37254653],
[-3.28707652, -1.84860763]])Covariance matrix
pca.get_covariance()array([[ 4.00000000e+00, -1.00000000e+00, 3.62064830e-15,
-1.00000000e+00, 7.00000000e+00],
[-1.00000000e+00, 3.33333333e-01, -5.00000000e-01,
6.66666667e-01, -2.00000000e+00],
[ 3.82368329e-15, -5.00000000e-01, 3.00000000e+00,
-2.50000000e+00, 1.50000000e+00],
[-1.00000000e+00, 6.66666667e-01, -2.50000000e+00,
2.33333333e+00, -3.00000000e+00],
[ 7.00000000e+00, -2.00000000e+00, 1.50000000e+00,
-3.00000000e+00, 1.30000000e+01]])
Using PySpark
As discussed before, we are using large datasets. Scikit-learn will crash on single computers trying to compute PCA on datasets such as these. As a result, we look to PySpark to distribute the computation of PCA.
Method 1: Spark's ML Package
The code below shows PCA in PySpark using Spark's ML package. The transformed matrix looks different than sklearn's result. This is because sklearn subtracts the mean of the input to make sure that the output is zero mean. However, the PCA module in PySpark applies transformation to the original input. Basically, sklearn computes $\bZ_{zeromean}$ (equation \ref{eqn:Zzeromean}) and PySpark computes $\bZ$ (equation \ref{eqn:Z}).
import os
import sys
# Path for Spark Source folder
os.environ['SPARK_HOME'] = '/Applications/spark-1.6.0-bin-hadoop2.6'
# Append pyspark to Python Path
sys.path.append('/Applications/spark-1.6.0-bin-hadoop2.6/python')
from pyspark import SparkContext
from pyspark import SparkConf
from pyspark.sql import SQLContextUsing Spark's PCA module in ML
from pyspark.ml.feature import PCA
from pyspark.mllib.linalg import Vectors
sc = SparkContext('local')
sqlContext = SQLContext(sc)
data = [(Vectors.dense([0.0, 1.0, 0.0, 7.0, 0.0]),),
Vectors.dense([2.0, 0.0, 3.0, 4.0, 5.0,]),
Vectors.dense([4.0, 0.0, 0.0, 6.0, 7.0])]
df = sqlContext.createDataFrame(data, ["features"])
pca_extracted = PCA(K=2, inputCol="features", outputCol="pca_features")
model = pca.extracted.fit(df)
features = model.transform(df) # this creates a DataFrame with the regular features and pca_featuresRepresentation in 2-D (matrix Z)
features.select("pca_features").collect()[Row(pca_features=DenseVector([1.6486, -4.0133])), Row(pca_features=DenseVector([-4.6451, -1.1168])), Row(pca_features=DenseVector([-6.4289, -5.338]))]Components (matrix V)
indetity_input = [(Vectors.dense([1.0, .0, 0.0, 0.0]),),
(Vectors.dense([.0, 1.0, .0, .0. .0]),),
(Vectors.dense([.0, 0.0, 1.0, .0, .0]),)
(Vectors.dense([.0, 0.0, .0, 1.0, .0]),)
(Vectors.dense([.0, 0.0, .0, .0, 1.0]),)]
df_identity = sqlContext.createDataFrame(identity_input, ['features'])
identity_features = model.transform(df_identity)
identity_features.select("pca_features").collect()[Row(pca_features=DenseVector([-0.4486, -0.2842])),Row(pca_features=DenseVector([0.133, -0.0562])),Row(pca_features=DenseVector([-0.1252, 0.7636])),Row(pca_features=DenseVector([0.2165, -0.5653])),Row(pca_features=DenseVector([-0.8477, -0.1156]))]You might have noticed that an identity matrix was necessary to pull out the features. Spark's PCA module had limited outputs, so to be able to see the components we had to use a dummy transformation on the features.
Method 2: SVD through Wrapping Java/Scala
In Spark 1.6.0 we can compute SVD using some modules from Spark's mllib package. However, SVD in Spark was implemented in Scala and Java. We resorted to making PySpark wrappers to those SVD classes using the functions written by Elias Abou Haydar.[1]
from pyspark.mllib.common import callMLlibFunc, JavaModelWrapper
from pyspark.mllib.linalg.distributed import RowMatrix
from pyspark.mllib.linalg import _convert_to_vector, Matrix, DenseMatrix
from pyspark.ml.feature import StandardScaler
class RowMatrix_new(RowMatrix):
def multiply(self, matrix):
"""
Multiplies the given RowMatrix with another matrix.
:param matrix: Matrix to multiply with.
:returns: RowMatrix
>>> rm = RowMatrix(sc.parallelize([[0, 1], [2, 3]]))
>>> rm.multiply(DenseMatrix(2, 2, [0, 2, 1, 3])).rows.collect()
[DenseVector([2.0, 3.0]), DenseVector([6.0, 11.0])]
"""
if not isinstance(matrix, DenseMatrix):
raise ValueError("Only multiplication with DenseMatrix "
"is supported.")
j_model = self._java_matrix_wrapper.call("multiply", matrix)
return RowMatrix_new(j_model)
class SVD(JavaModelWrapper):
"""Wrapper around the SVD scala case class"""
@property
def U(self):
""" Returns a RowMatrix whose columns are the left singular vectors of the SVD if computeU was set to be True."""
u = self.call("U")
if u is not None:
return RowMatrix(u)
@property
def s(self):
"""Returns a DenseVector with singular values in descending order."""
return self.call("s")
@property
def V(self):
""" Returns a DenseMatrix whose columns are the right singular vectors of the SVD."""
return self.call("V")
def computeSVD(row_matrix, k, computeU=False, rCond=1e-9):
"""
Computes the singular value decomposition of the RowMatrix.
The given row matrix A of dimension (m X n) is decomposed into U * s * V'T where
* s: DenseVector consisting of square root of the eigenvalues (singular values) in descending order.
* U: (m X k) (left singular vectors) is a RowMatrix whose columns are the eigenvectors of (A X A')
* v: (n X k) (right singular vectors) is a Matrix whose columns are the eigenvectors of (A' X A)
:param k: number of singular values to keep. We might return less than k if there are numerically zero singular values.
:param computeU: Whether of not to compute U. If set to be True, then U is computed by A * V * sigma^-1
:param rCond: the reciprocal condition number. All singular values smaller than rCond * sigma(0) are treated as zero, where sigma(0) is the largest singular value.
:returns: SVD object
"""
java_model = row_matrix._java_matrix_wrapper.call("computeSVD", int(k), computeU, float(rCond))
return SVD(java_model)
def computePCA_sergul(input, numReducedDim):
"""
Computer PCA for a given input using SVD
:param input: A matrix of dimension num_of_observation x num_of_features
:param numReducedDim: Dimension of reduced space
:return: components, projection and singular values
"""
df = sqlContext.createDataFrame(input,["features"])
standardizer = StandardScaler(withMean=True, withStd=False,
inputCol='features',
outputCol='std_features')
model_normalizer = standardizer.fit(df)
df_normalized = model_normalizer.transform(df)
pca_features = df_normalized.select("std_features").rdd.map(lambda row : row[0])
mat = RowMatrix_new(pca_features)
svd = computeSVD(mat,numReducedDim,True)
components = svd.call("V").toArray() # dim_of_data x reduced dim
projection = RowMatrix_new((pca_features)).multiply(svd.call("V"))
singularValues = svd.call("s").toArray()
return components, projection, singularValues
\end{lstlisting}We can apply this framework to the same input from the toy example as shown below.
data = sc.parallelize([(Vectors.dense([0.0, 1.0, 0.0, 7.0, 0.0]),),
Vectors.dense([2.0, 0.0, 3.0, 4.0, 5.0,]),
Vectors.dense([4.0, 0.0, 0.0, 6.0, 7.0])])
components, projection, singularValues = computePCA_sergul(input=data,
numReducedDim=2)Components (matrix V)
componentsarray([[-0.44859172, -0.28423808],[0.13301986, -0.5621156],[-0.12523156, 0.76362648], [0.21650757, -0.56529588], [-0.84765129, -0.11560341]])Representation in 2-D, (matrix Z)
projection.rows.collect()[DenseVector([4.7904, -0.5239]), DenseVector([-1.5033, 2.3725]), DenseVector([-3.2871, -1.8486])]Singular values (matrix S)
singularValues()array([6.00104109, 3.05300494])The results are consistent with the theoretical results in section 2.
Application: Functional magnetic resonance imaging
We applied PCA to a neuroimaging data set to explore neuronal signatures in the human brain. We used resting state functional magnetic resonance imaging data (fMRI) for 900 subjects, collected as part of the Human Connectome Project (HCP).[3]
The preprocessing of this data set is beyond the scope of this article, but you can see the dimensions involved in creating the dataset in Figure 1.
The number of time points for each subject is 1,200 and the number of voxels is 13,000. Since there are 900 subjects, we need to compute SVD on matrix of size $1080000 \times 13000$ (matrix $\bX^T$ in our notation).
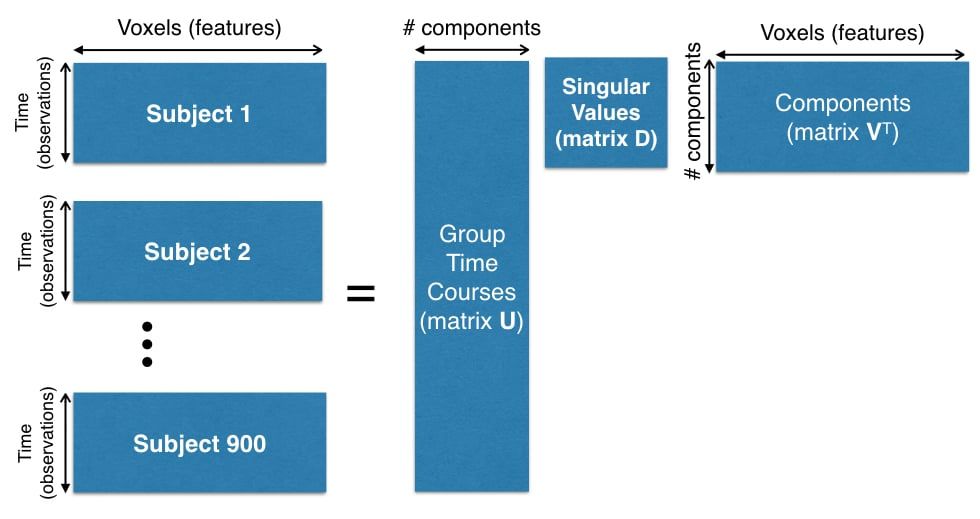
Figure 1. Concatenation of 900 subjects for PCA computation.
Computing SVD on a matrix of size $1080000 \times 13000$ is computationally expensive. Therefore, we backed PySpark with commodity hardware provided by Amazon Web Services and accessed through the Domino data science platform.
PCA on the entire dataset took 27 hours using hardware with 16 cores and 30GB RAM. Figure 2 shows the top four components from four different views.
While interpreting such images is hard, two conclusions are of note:
1. The maps are roughly symmetric, in line with what is expected for resting state networks of the brain.
2. The third component has hotspots at the dorsal medial prefrontal cortex and the precuneus, two key components of the default mode network.
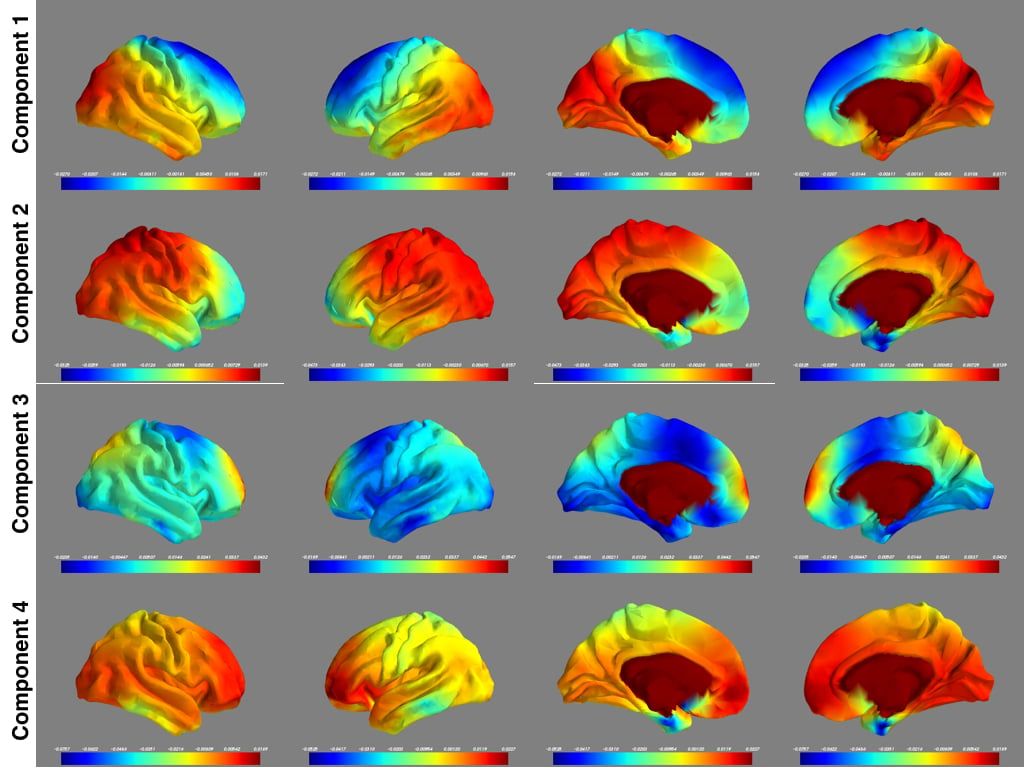
Figure 2. Concatenation of 900 subjects for PCA computation.
References
- Elias Abou Haydar. http://stackoverflow.com/a/33500704/4544800. 2015.
- Jonathon Shlens. A tutorial on principal component analysis. arXiv preprint
arXiv:1404.1100, 2014. - Stephen M Smith, Christian F Beckmann, Jesper Andersson, Edward J Auerbach, Janine Bijsterbosch, Gwenaëlle Douaud, Eugene Duff, David A Feinberg, Ludovica Griffanti, Michael P Harms, et al. Resting-state fmri in the human connectome project. Neuroimage, 80:144–168, 2013.
Banner image titled "DSC_2236" by Edward Blake. Licensed under CC BY 2.0
LaTeX rendering by QuickLaTeX.
Sergul and Syed received their Ph.D.s in Electrical Engineering in 2014 from the University of Southern California, applying signal processing to neuroimaging data. They continue to use machine learning on brain imaging data as a pastime and sharing their knowledge with the community. They constantly challenge each other as good buddies and like to call themselves “signal learners.”


