Python 3 Support in Jupyter
Jonathan Schaller2016-01-12 | 4 min read
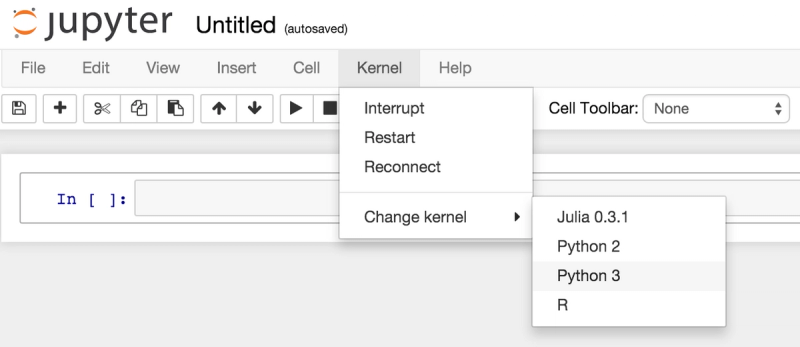
Domino lets you spin up Jupyter notebooks (and other interactive tools) with one click, on powerful cloud hardware. We recently added beta support for Python 3, so you can now run Jupyter Notebooks in Python 2 or 3:
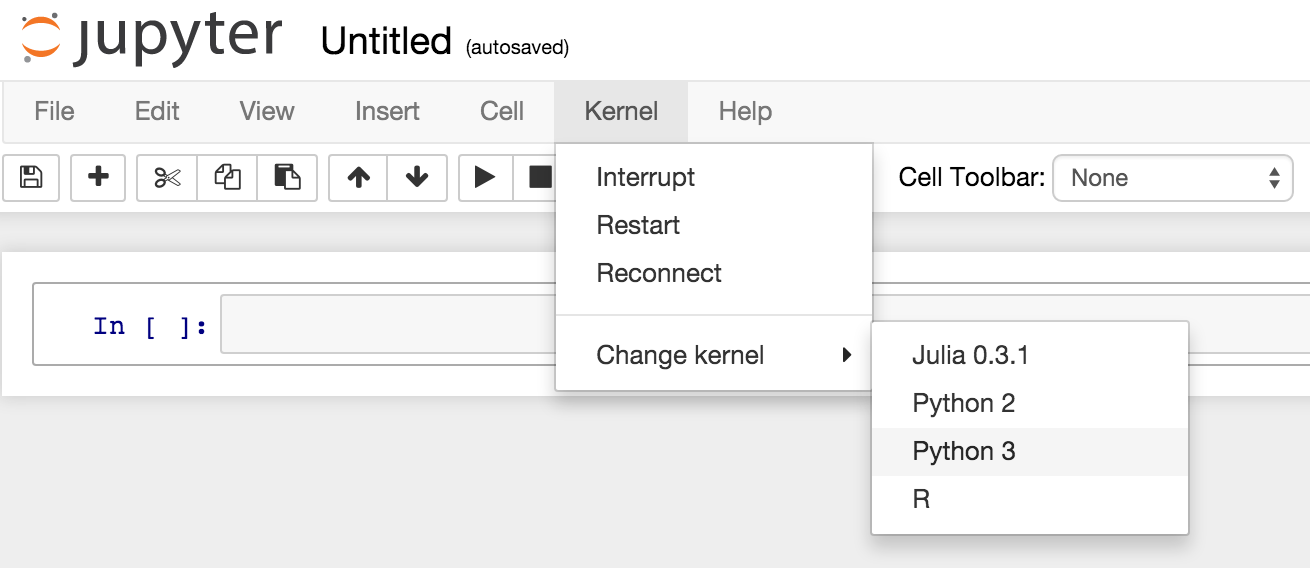
So if you’ve been wanting to try Python 3, but haven’t wanted to deal with maintaining an installation on your machine, you can give it a shot on Domino. Please contact us if you're interested and we'll give you access to this beta feature.
What's new in Python 3
Python 3 includes a number of syntax improvements and other features. For example:
- Better unicode handling (all strings are unicode by defaults), which can be handy for NLP projects
- Function annotations
- Dictionary comprehensions
- Many common APIs return iterators instead of lists, which can guard against OOM errors
- And more
Installation & Setup
For the curious, here’s a guide to how I got Python 3 to run alongside Python 2 in Jupyter. While it's straightforward to set up a standalone Python 3 Jupyter kernel, supporting Python 2 and 3 simultaneously turned out to be trickier than anticipated. I didn't find a clear guide for setting it up this way, so I wanted to pass along my learnings.
The installation involves a few components:
- Jupyter needs to be set up to utilize both Python 2 and 3. This involves installing a few prerequisite dependencies, and making sure the kernelspecs for both Python versions are available to Jupyter on the filesystem.
- Since Python 3 is independent from any existing Python 2 installation, it's necessary to set up a separate package manager (pip3). This is used to install additional IPython dependencies as well as some common scientific libraries.
- Lastly, a few bugs arise due to the installation that need to be cleaned up.
Here are the commands I used. Notes and additional details follow:
apt-get install python3-setuptools python3-dev libzmq-dev
easy_install3 pip# More dependencies needed to run Python 3 in Jupyterpip3 install ipython==3.2.1 pyzmq jinja2 tornado jsonschema# IPython kernelspecs
ipython3 kernelspec install-self
ipython2 kernelspec install-self# Install some libraries
pip3 install numpy scipy scikit-learn pandas matplotlib# Bug cleanup:# Fix Jupyter terminals by switching IPython back to use Python 2
sed -i.bak 's/python3/python/' /usr/local/bin/ipython# Reset the "default" pip to pip2, if desired
pip2 install --upgrade --force-reinstall pip# Fix a link broken by python3-dev that led to errors when running R
echo | update-alternatives --config libblas.so.3# Make sure the local site packages dir exists mkdir -p ~/.local/lib/python3.4/site-packages
Notes on the installation:
- While running a notebook, you can add new packages interactively with
! pip3 install --user <package==version>, and check which packages are already installed by running! pip3 freeze. If you need additional packages, but interactive runtime installation is not ideal, please let us know and we can help set you up with a custom environment. - Kernelspec installation commands come from the IPython docs.
- After installing both Python kernelspecs, Jupyter mostly works fine, except for terminals, which are listed as “unavailable” in the session. I tracked the bug down to a dependency issue. After running the preceding commands, IPython is run by Python 3: the initial shebang line of
/usr/local/bin/ipythonreads#!/usr/bin/python3, and the Python 3 installation can't find a module needed to run Jupyter terminals. Rather than trying to fix that, it was easier to just tell IPython to use Python 2 again, by editing the initial line to read#!/usr/bin/pythonwith thesedcommand. That works, and terminals are back online. - Installing pip3 causes the
pipcommand to runpip3instead ofpip2. Thepip2 install --upgrade --force-reinstall pipcommand revertspipback topip2, which is what we wanted so that Python 2 remains the "default" Python version. - The
update-alternatives --config libblas.so.3command fixes a broken link introduced byapt-get install python3-dev. Without this command, R scripts produce the following error:
Error in dyn.load(file, DLLpath = DLLpath, ...) :
unable to load shared object '/usr/lib/R/library/stats/libs/stats.so':
/usr/lib/liblapack.so.3: undefined symbol: ATL_chemv
During startup - Warning message:
package 'stats' in options("defaultPackages") was not found- A final issue cropped up: pip3's local installation directory (in this case,
~/.local/lib/python3.4/site-packages/) wasn't on Python 3'ssys.path. It turns out that Python'ssitemodule (which is supposed to add this path tosys.pathwhen Python is started) was ignoring the path because it did not exist yet. Creating this directory ahead of time solves the problem. Note that the user running Jupyter must have read/write access to this directory.)
Questions, concerns, or just want to test out this environment? Contact us!


