R Notebooks in the Cloud
Nick Elprin2014-10-11 | 5 min read

We recently added a feature to Domino that lets you spin up an interactive R session on any class of hardware you choose, with a single click, enabling more powerful interactive, exploratory work in R without any infrastructure or setup hassle. This post describes how and why we built our "R Notebook" feature.
How R Notebooks work
We wanted a solution that: (1) let our users work with R interactively; (2) on powerful machines; and (3) run R on the cloud without requiring any setup or infrastructure management. For reasons I describe below, we adapted IPython Notebook to fill this need. The result is what we call an R Notebook: an interactive, IPython Notebook environment that works with R code. It even handles plotting and visual output!
So how does it work?
Step 1: Start a notebook session with one click:
Like any other run in Domino, this will spin up a new machine (on hardware of your choosing), and automatically load it with your project files.
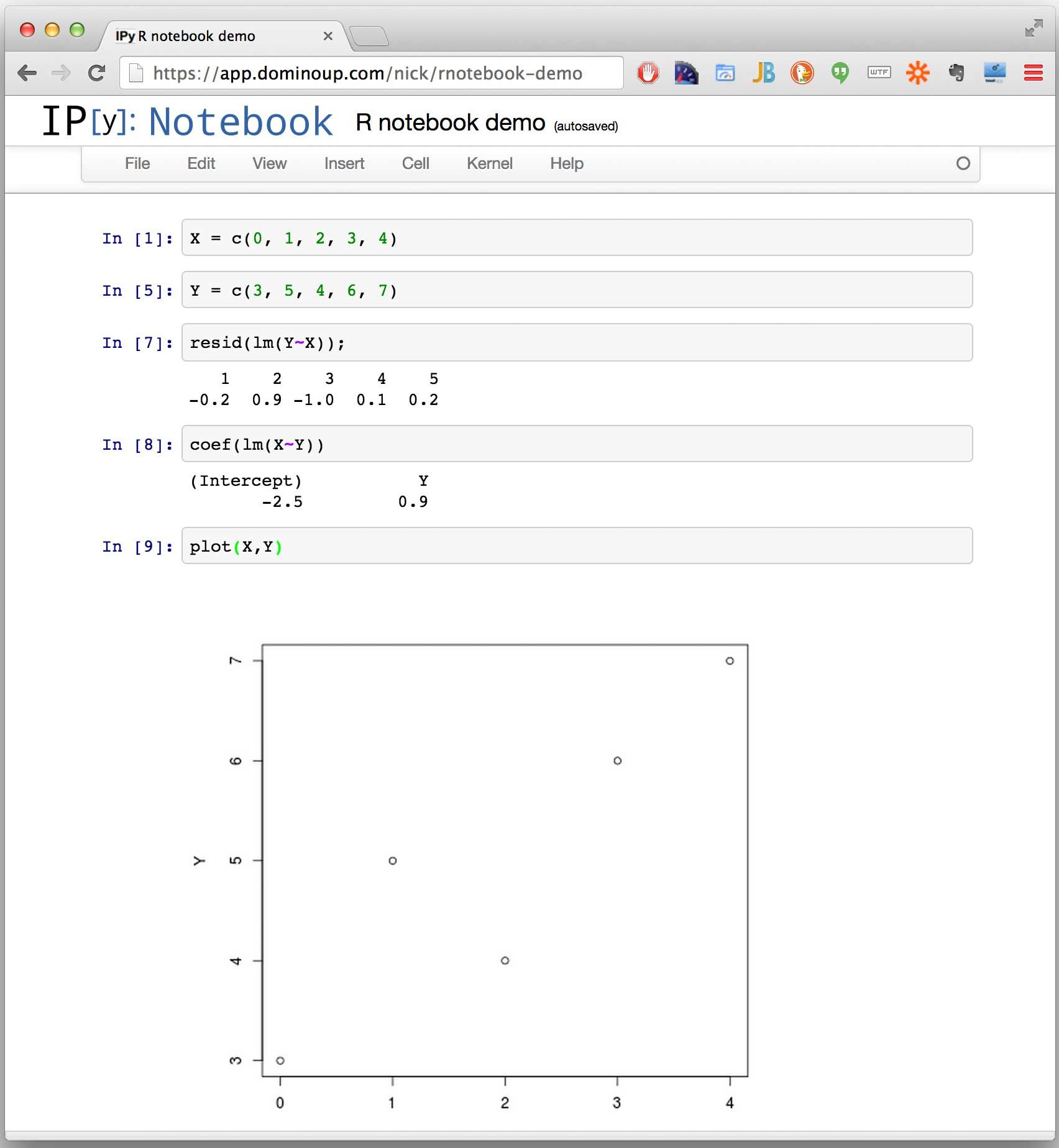
Step 2: Use the notebook!
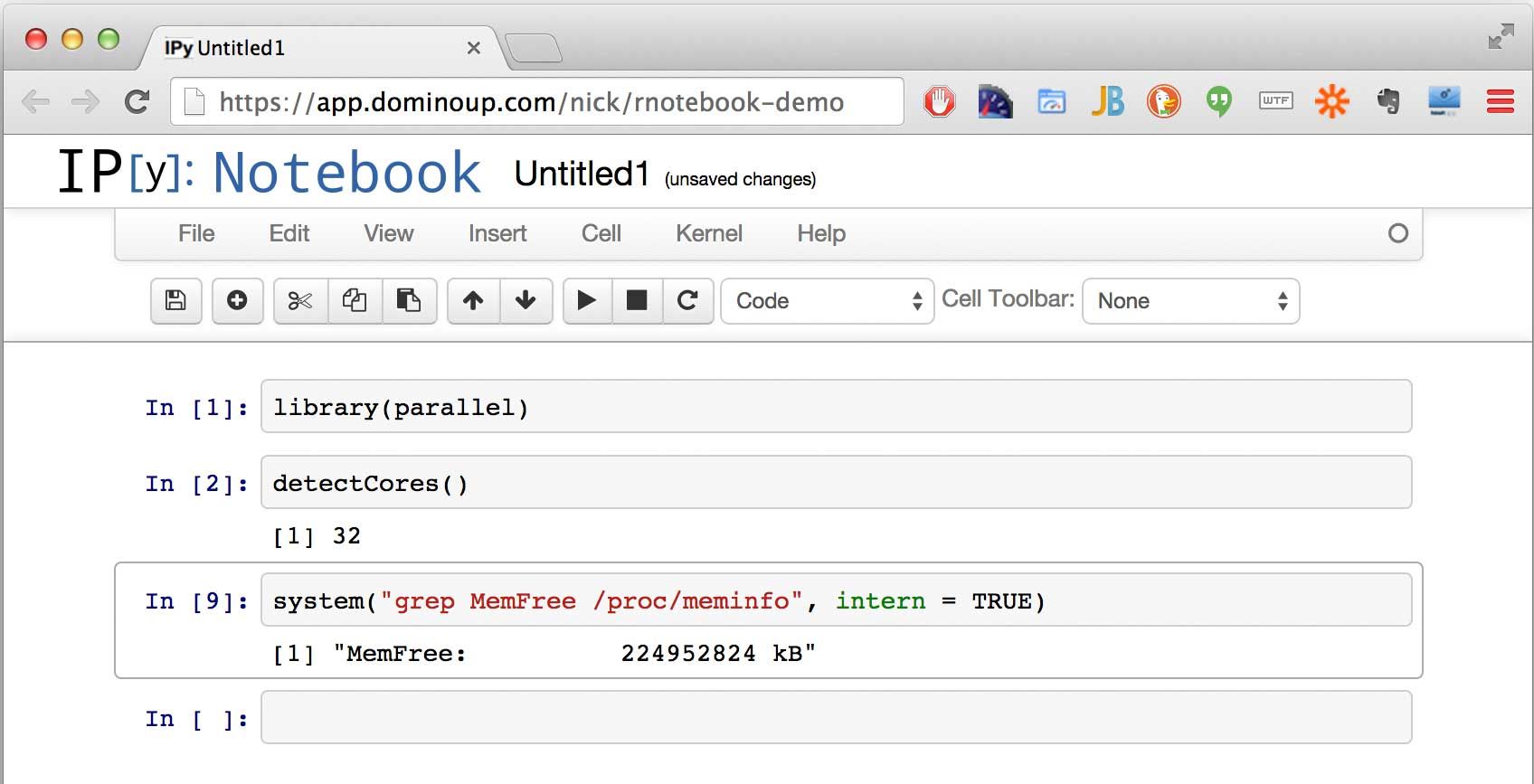
Any R command will work, including ones that load packages, and the system function. Since Domino lets you spin up these notebooks on ridiculously powerful machines (e.g., 32 cores, 240GB of memory), let's show off a bit:
Motivation
Our vision for Domino is to be a platform that accelerates work across the entire analytical lifecycle, from early exploration, all the way to packaging and deployment of analytical models. We think we're well on our way toward that goal, and this post is about a recent feature we added to fill a gap in our support for early stages of that lifecycle: interactive work in R.
The Analytical Lifecycle
Analytical ideas move through different phases:
- Exploration / Ideation. In the early stages of an idea, it's critical to be able to "play with data" interactively. You are trying different techniques, fixing issues quickly, to figure out what might work.
- Refinement. Eventually you have an approach that you want to invest in, and you must refine or "harden" a model. Often this requires many more intensive experiments: for example, running a model over your entire data set with several different parameters, to see what works best.
- Packaging and Deployment. Once you have something that works, typically it will be deployed for some ongoing use: either packaged into a UI for people to interact with, or deployed with some API (or web service) so software systems can consume it.
Domino offers solutions for all three phases, in multiple different languages, but we had a gap. For interactive exploratory work, we support IPython Notebooks for work in Python, but we didn't have a good solution for work in R.
Interactive environment | Able to run many experiments in parallel, quickly, and track work and results | Easily create a GUI or web service around your model |
|---|---|---|
Our | Our bread and butter: easily run your scripts on remote machines, as many as you want, and keep them all tracked | Launchers for UI, and RServe powering API publishing |
IPython Notebooks | Launchers for UI, and pyro powering API publishing |
Implementation details
Since we already had support for spinning up IPython Notebook servers inside docker containers on arbitrary EC2 machines, we opted to use IPython Notebook for our R solution.
A little-known fact about IPython Notebook (likely because of its name) is that it can actually run code in a variety of other languages. In particular, its RMagic functionality lets you run R commands inside IPython Notebook cells by prepending your commands with the %R modifier. We adapted this "hack" (thanks, fperez!) to prepend the RMagic modifying automatically to every cell expression.
The approach is to make a new ipython profile with a startup script that automatically prepends the %R magic prefix to any expression you evaluate. The result is an interactive R notebook.
The exact steps were:
- pip install rpy2
- ipython profile create rkernel
- Copy rkernel.py into ~/.ipython/profile_rkernel/startup
Where rkernely.py is a slightly-modified version of fperez's script. We just had to change the rmagic extension on line 15 to the rpy2.ipython extension, to be compatible with IPython Notebook 2.
A "native" IPython R kernel in 15 lines of code.
This isn't a real native R kernel, just a quick and dirty hack to get the basics running in a few lines of code.
Put this into your startup directory for a profile named 'rkernel' or somesuch, and upon startup, the kernel will imitate an R one by simply prepending %%R to every cell.
from IPython.core.interactiveshell import InteractiveShell
print('*** Initializing R Kernel ***')
ip = get_ipython()
ip.run_line_magic('load_ext', 'rpy2.ipython')
ip.run_line_magic('config', 'Application.verbose_crash=True')
old_run_cell = InteractiveShell.run_cell
def run_cell(self, raw_cell, **kw):
return old_run_cell(self, '%%Rn' + raw_cell, **kw)
InteractiveShell.run_cell = run_cellWhat about RStudio server?
Some folks who have used this have asked why we didn't just integrate RStudio Server, so you could spin up an RStudio session in the browser. The honest answer is that using IPython Notebook was much easier, since we already supported it. We are exploring an integration with RStudio Server, though. Please let us know if you would use it.
In the meantime, try out the R Notebook functionality!
Nick Elprin is the CEO and co-founder of Domino Data Lab, provider of the open data science platform that powers model-driven enterprises such as Allstate, Bristol Myers Squibb, Dell and Lockheed Martin. Before starting Domino, Nick built tools for quantitative researchers at Bridgewater, one of the world's largest hedge funds. He has over a decade of experience working with data scientists at advanced enterprises. He holds a BA and MS in computer science from Harvard.



