Python for SAS users: the Pandas data analysis library
Randy Betancourt2016-12-20 | 15 min read

This post is a chapter from Randy Betancourt's Python for SAS Users quick start guide. Randy wrote this guide to familiarize SAS users with Python and Python's various scientific computing tools.
An introduction to Pandas
This chapter introduces the pandas library (or package). pandas provides Python developers with high-performance, easy-to-use data structures and data analysis tools. The package is built on NumPy (pronounced 'numb pie'), a foundational scientific computing package that offers the ndarray, a performant object for array arithmetic. We will illustrate a few useful NumPy objects as a way of illustrating pandas.
For data analysis tasks we often need to group dissimilar data types together. An example being grouping categorical data using strings with frequencies and counts using ints and floats for continuous values. In addition, we would like to be able to attach labels to columns, pivot data, and so on.
We begin by introducing the Series object as a component of the DataFrame object. A Series can be thought of as an indexed, one-dimensional array, similar to a column of values. DataFrames can be thought of as a two-dimensional array indexed by both rows and columns. A good analogy is an Excel cell addressable by row and column location.
In other words, a DataFrame looks a great deal like a SAS data set (or relational table). The table below compares pandas components to those found in SAS.
DataFrame | SAS data set |
|---|---|
row | observation |
column | variable |
groupby | BY-Group |
NaN | . |
slice | sub-set |
axis 0 | observation |
axis 1 | column |
DataFrame and Series indexes are covered in detail in Chapter 6, Understanding Indexes.
Importing packages
To begin utilizing pandas objects, or objects from any other Python package, we begin by importing libraries by name into our namespace. To avoid having to retype full package names repeatedly, use the standard aliases of np for NumPy and pd for pandas.
import numpy as np
import pandas as pd
from numpy.random import randn
from pandas import Series, DataFrame, Index
from IPython.display import ImageSeries
A Series can be thought of as a one-dimensional array with labels. This structure includes an index of labels used as keys to locate values. Data in a Series can be of any data type. pandas data types are covered in detail here . In the SAS examples, we use Data Step ARRAYs as an analog to the Series.
Start by creating a Series of random values:
s1 = Series(randn(10))
print(s1.head(5))0 -0.467231
1 -0.504621
2 -0.122834
3 -0.418523
4 -0.262280
dtype: float64Notice the index start position begins with 0. Most SAS automatic variables like _n_ use 1 as the index start position. Iteration of the SAS DO loop 0 to 9 in conjunction with an ARRAY produces an array subscript out of range error.
In the SAS example below the DO loop is used to iterate over the array elements locating the target elements.
Arrays in SAS are used primarily for iteratively processing like variables together. SAS/IML is a closer analog to NumPy arrays. SAS/IML is outside the scope of these examples.
0.4322317772
0.5977982976
0.7785986473
0.1748250183
0.3941470125A Series can have a list of index labels.
s2 = Series(randn(10), index=["a", "b", "c", "d", "e", "f", "g", "h", "i", "j"])
print(s2.head(5))
a -1.253542
b 1.093102
c -1.248273
d -0.549739
e 0.557109
dtype: float64The Series is indexed by integer value with the start position at 0.
print(s2[0])-1.25354189867The SAS example uses a DO loop as the index subscript into the array.
0.4322317772Return the first 3 elements in the Series.
print(s2[:3])a -1.253542
b 1.093102
c -1.248273
dtype: float640.4322317772
0.5977982976
0.7785986473The example has two operations. The s2.mean() method calculates mean followed by a boolean test less than this calculated mean.
s2[s2 < s2.mean()]a -1.253542
c -1.248273
d -0.549739
h -2.866764
i -1.692353
dtype: float64Series and other objects have attributes that use a dot (.) chaining-style syntax. .name is one of a number of attributes for the Series object.
s2.name="Arbitrary Name"
print(s2.head(5))
a -1.253542
b 1.093102
c -1.248273
d -0.549739
e 0.557109
Name: Arbitrary Name, dtype: float64DataFrames
As stated earlier, DataFrames are relational-like structures with labels. Alternatively, a DataFrame with a single column is a Series.
Like SAS, DataFrames have different methods for creation. DataFrames can be created by loading values from other Python objects. Data values can also be loaded from a range of non-Python input sources, including .csv files, DBMS tables, Web API's, and even SAS data sets (.sas7bdat), etc. Details are discussed in Chapter 11 -- pandas Readers .
Start by reading the UK_Accidents.csv file. It contains vehicular accident data in the U.K from January 1, 2015 to December 31, 2015. The .csv file is located here.
There are multiple reports for each day of the year, with values being mostly integers. Another .CSV file found here maps values to descriptive labels.
Read .csv files
The default values are used in the example below. pandas provides a number of readers with parameters for controlling missing values, date parsing, line skipping, data type mapping, etc. These parameters are analogous to SAS' INFILE/INPUT processing.
Notice the additional backslash \\ to normalize the Window's path name.
file_loc2 = "C:\Data\uk_accidents.csv"
df = pd.read_csv(file_loc2, low_memory=False)PROC IMPORT is used to read the same .csv file. This is one of several methods for SAS to read a .csv file. Here we have taken the defaults.
NOTE: The file 'c:\data\uk_accidents.csv' is:
File Name 'c:\data\uk_accidents.csv',
Lrecl=32760, Recfm=VNOTE: 266776 records were read from file 'c:\data\uk_accidents.csv' The minimum record length was 65 The maximum record length was 77 NOTE: Data set "WORK.uk_accidents" has 266776 observation(s) and 27 variable(s)Unlike SAS, the Python interpreter is mainly silent upon normal execution. When debugging it is helpful to invoke methods and functions to return information about these objects. This is somewhat analogous to use PUT statements in the SAS log to examine variable values.
The size, shape, and ndim attributes (respectively, number of cells, rows/columns, and number of dimensions) are shown below.
print(df.size, df.shape, df.ndim)7202952 (266776, 27) 2Read verification
After reading a file, you often want to understand its content and structure. The DataFrame .info() method returns descriptions of the DataFrame's attributes.
df.info()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 266776 entries, 0 to 266775
Data columns (total 27 columns):
Accident_Severity 266776 non-null int64
Number_of_Vehicles 266776 non-null int64
Number_of_Casualties 266776 non-null int64
Day_of_Week 266776 non-null int64
Time 266752 non-null object
Road_Type 266776 non-null int64
Speed_limit 266776 non-null int64
Junction_Detail 266776 non-null int64
Light_Conditions 266776 non-null int64
Weather_Conditions 266776 non-null int64
Road_Surface_Conditions 266776 non-null int64
Urban_or_Rural_Area 266776 non-null int64
Vehicle_Reference 266776 non-null int64
Vehicle_Type 266776 non-null int64
Skidding_and_Overturning 266776 non-null int64
Was_Vehicle_Left_Hand_Drive_ 266776 non-null int64
Sex_of_Driver 266776 non-null int64
Age_of_Driver 266776 non-null int64
Engine_Capacity__CC_ 266776 non-null int64
Propulsion_Code 266776 non-null int64
Age_of_Vehicle 266776 non-null int64
Casualty_Class 266776 non-null int64
Sex_of_Casualty 266776 non-null int64
Age_of_Casualty 266776 non-null int64
Casualty_Severity 266776 non-null int64
Car_Passenger 266776 non-null int64
Date 266776 non-null object
dtypes: int64(25), object(2)
memory usage: 55.0+ MBIn SAS, this same information is generally found in the output from PROC CONTENTS.
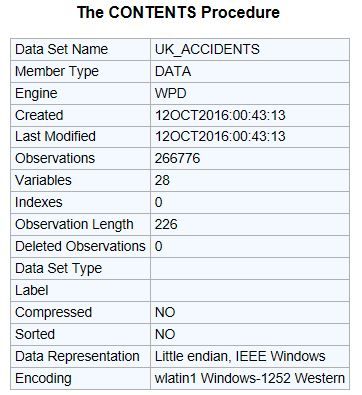

Inspection
pandas has methods useful for inspecting data values. The DataFrame .head() method displays the first 5 rows by default. The .tail() method displays the last 5 rows by default. The row count value can be an arbitrary integer value such as:
# display the last 20 rows of the DataFrame
df.tail(20)SAS uses the FIRSTOBS and OBS options with procedures to determine input observations. The SAS code to print the last 20 observations of the uk_accidents data set is:
df.head()5 rows × 27 columnsOBS=n in SAS determines the number of observations used as input.
The output from PROC PRINT is not displayed here.Scoping output by columns is shown in the cell below. The column list is analogous to the VAR statement in PROC PRINT. Note the double set of square brackets for this syntax. This example illustrates slicing by column label. Slicers work along rows as well. The square braces [] are the slicing operator. The details are explained here
df[["Sex_of_Driver", "Time"]].head(10)1 | 19:00 |
|---|---|
1 | 19:00 |
1 | 18:30 |
2 | 18:30 |
1 | 18:30 |
1 | 17:50 |
1 | 17:50 |
1 | 7:05 |
1 | 7:05 |
1 | 12:30 |
Notice the DataFrame default index (incrementing from 0 to 9). This is analogous to the SAS automatic variable n. Later, we illustrate using other columns in the DataFrame as the index.
Below is the SAS program to print the first 10 observations of a data set along with the variables Sec_of_Driver and Time.
The output from PROC PRINT is not displayed here.Handling missing data
Before analyzing data, a common task is dealing with missing data. pandas uses two designations to indicate missing data, NaN (not a number) and the Python None object.
The cell below uses the Python None object to represent a missing value in the array. In turn, Python infers the data type for the array to be an object. Unfortunately, the use of a Python None object with an aggregation function for arrays raises an error.
s1 = np.array([32, None, 17, 109, 201])
s1array([32, None, 17, 109, 201], dtype=object)s1.sum()---------------------------------------------------------------------------TypeError Traceback (most recent call last) in <module>----> 1 s1.sum()/opt/anaconda3/envs/reporting_env/lib/python3.8/site-packages/numpy/core/_methods.py in _sum(a, axis, dtype, out, keepdims, initial, where) 45 def _sum(a, axis=None, dtype=None, out=None, keepdims=False, 46 initial=_NoValue, where=True):---> 47 return umr_sum(a, axis, dtype, out, keepdims, initial, where) 48 49 def _prod(a, axis=None, dtype=None, out=None, keepdims=False,TypeError: unsupported operand type(s) for +: 'int' and 'NoneType'To alleviate the error raised above, use the np.nan (missing value indicator) in the array example below. Also notice how Python chose floating point (or up-casting) for the array compared to the same example two cells above.
s1 = np.array([32, np.nan, 17, 109, 201])
print(s1)
s1.dtype[ 32. nan 17. 109. 201.]dtype('float64')
Not all arithmetic operations using NaN's will result in a NaN.
s1.mean()nanContrast the Python program in the cell above for calculating the mean of the array elements with the SAS example below. SAS excludes the missing value and utilizes the remaining array elements to calculate a mean.
89.75Missing value identification
Returning to our DataFrame, we need an analysis of missing values for all the columns. Pandas provide four methods for the detection and replacement of missing values. They are:
isnull()generates a boolean mask to indicate missing values
Method | Action Taken |
|---|---|
| opposite of |
| returns a filtered version of the data |
| returns a copy of data with missing values filled or imputed |
We will look at each of these in detail below.
A typical SAS-programming approach to address the missing data analysis is to write a program to traverses all columns using counter variables with IF/THEN testing for missing values.
This can be along the lines of the example in the output cell below. df.columns returns the sequence of column names in the DataFrame.
for col_name in df.columns:
print(col_name, end="---->")
print(sum(df[col_name].isnull()))Accident_Severity---->0
Number_of_Vehicles---->0
Number_of_Casualties---->0
Day_of_Week---->0
Time---->24
Road_Type---->0
Speed_limit---->0
Junction_Detail---->0
Light_Conditions---->0
Weather_Conditions---->0
Road_Surface_Conditions---->0
Urban_or_Rural_Area---->0
Vehicle_Reference---->0
Vehicle_Type---->0
Skidding_and_Overturning---->0
Was_Vehicle_Left_Hand_Drive_---->0
Sex_of_Driver---->0
Age_of_Driver---->0
Engine_Capacity__CC_---->0
Propulsion_Code---->0
Age_of_Vehicle---->0
Casualty_Class---->0
Sex_of_Casualty---->0
Age_of_Casualty---->0
Casualty_Severity---->0
Car_Passenger---->0
Date---->0While this give the desired results, there is a better approach.
As an aside, if you find yourself thinking of solving a pandas operation (or Python for that matter) using iterative processing, stop and take a little time to do research. Chances are, a method or function already exists!
Case-in-point is illustrated below. It chains the .sum() attribute to the .isnull() attribute to return a count of the missing values for the columns in the DataFrame.
The .isnull() method returns True for missing values. By chaining the .sum() method to the .isnull() method it produces a count of the missing values for each columns.
df.isnull().sum()Accident_Severity 0
Number_of_Vehicles 0
Number_of_Casualties 0
Day_of_Week 0
Time 24
Road_Type 0
Speed_limit 0
Junction_Detail 0
Light_Conditions 0
Weather_Conditions 0
Road_Surface_Conditions 0
Urban_or_Rural_Area 0
Vehicle_Reference 0
Vehicle_Type 0
Skidding_and_Overturning 0
Was_Vehicle_Left_Hand_Drive_ 0
Sex_of_Driver 0
Age_of_Driver 0
Engine_Capacity__CC_ 0
Propulsion_Code 0
Age_of_Vehicle 0
Casualty_Class 0
Sex_of_Casualty 0
Age_of_Casualty 0
Casualty_Severity 0
Car_Passenger 0
Date 0
dtype: int64To identify missing values the SAS example below uses PROC Format to bin missing and non-missing values. Missing values are represented by default as (.) for numeric and blank (' ') for character variables. Therefore, a user-defined format is needed for both types.
PROC FREQ is used with the automatic variables _CHARACTER_ and _NUMERIC_ to produce a frequency listing for each variable type.
Only a portion of the SAS output is shown since separate output is produced for each variable. As with the Python for loop example above, the time variable is the only variable with missing values.

Another method for detecting missing values is to search column-wise by using the axis=1 parameter to the chained attributes .isnull().any(). The operation is then performed along columns.
null_data = df[df.isnull().any(axis=1)]null_data.head()5 rows × 27 columnsMissing value replacement
The code below is used to render multiple objects side-by-side. It is from Essential Tools for Working With Data, by Jake VanderPlas. It displays the 'before' and 'after' effects of changes to objects.
class display(object):
"""Display HTML representation of multiple objects"""
template = """<div style="float: left; padding: 10px;"><p style="font-family: 'Courier New', Courier, monospace;">{0}</p> {1}</div>"""
def __init__(self, *args):
self.args = args
def _repr_html_(self):
return "\n".join(self.template.format(a, eval(a)._repr_html_()) for a in self.args)
def __repr__(self):
return "\n".join(a + "" + repr(eval(a)) for a in self.args)To illustrate the .fillna() method, consider the following to create a DataFrame.
df2 = pd.DataFrame([["cold","slow", np.nan, 2., 6., 3.],
["warm", "medium", 4, 5, 7, 9],
["hot", "fast", 9, 4, np.nan, 6],
["cool", None, np.nan, np.nan, 17, 89],
['"cool","medium",, 16, 44, 21, 13],
["cold","slow", np.nan, 29, 33, 17]],
columns=["col1", "col2", "col3", "col4", "col5", "col6"],
index=(list('abcdef')))
display("df2")df2
cold | slow | NaN | 2.0 | 6.0 | 3.0 |
|---|---|---|---|---|---|
warm | medium | 4.0 | 5.0 | 7.0 | 9.0 |
hot | fast | 9.0 | 4.0 | NaN | 6.0 |
cool | None | NaN | NaN | 17.0 | 89.0 |
cool | medium | 16.0 | 44.0 | 21.0 | 13.0 |
cold | slow | NaN | 29.0 | 33.0 | 17.0 |
df_tf = df2.isnull()
display("df2", "df_tf")
df2
cold | slow | NaN | 2.0 | 6.0 | 3.0 |
|---|---|---|---|---|---|
warm | medium | 4.0 | 5.0 | 7.0 | 9.0 |
hot | fast | 9.0 | 4.0 | NaN | 6.0 |
cool | None | NaN | NaN | 17.0 | 89.0 |
cool | medium | 16.0 | 44.0 | 21.0 | 13.0 |
cold | slow | NaN | 29.0 | 33.0 | 17.0 |
False | False | True | False | False | False |
|---|---|---|---|---|---|
False | False | False | False | False | False |
False | False | False | False | True | False |
False | True | True | True | False | False |
False | False | False | False | False | False |
False | False | True | False | False | False |
By default the .dropna() method drops either the entire row or column in which any null value is found.
df3 = df2.dropna()display("df2", "df3")df2
cold | slow | NaN | 2.0 | 6.0 | 3.0 |
|---|---|---|---|---|---|
warm | medium | 4.0 | 5.0 | 7.0 | 9.0 |
hot | fast | 9.0 | 4.0 | NaN | 6.0 |
cool | None | NaN | NaN | 17.0 | 89.0 |
cool | medium | 16.0 | 44.0 | 21.0 | 13.0 |
cold | slow | NaN | 29.0 | 33.0 | 17.0 |
df3
warm | medium | 4.0 | 5.0 | 7.0 | 9.0 |
|---|---|---|---|---|---|
cool | medium | 16.0 | 44.0 | 21.0 | 13.0 |
The .dropna() method also works along a column axis. axis = 1 or axis = 'columns' is equivalent.
df4 = df2.dropna(axis="columns")
display("df2", "df4")df2
cold | slow | NaN | 2.0 | 6.0 | 3.0 |
|---|---|---|---|---|---|
warm | medium | 4.0 | 5.0 | 7.0 | 9.0 |
hot | fast | 9.0 | 4.0 | NaN | 6.0 |
cool | None | NaN | NaN | 17.0 | 89.0 |
cool | medium | 16.0 | 44.0 | 21.0 | 13.0 |
cold | slow | NaN | 29.0 | 33.0 | 17.0 |
df4
cold | 3.0 |
|---|---|
warm | 9.0 |
hot | 6.0 |
cool | 89.0 |
cool | 13.0 |
cold | 17.0 |
Clearly this drops a fair amount of 'good' data. The thresh parameter allows you to specify a minimum of non-null values to be kept for the row or column. In this case, row 'd' is dropped because it contains only 3 non-null values.
df5 = df2.dropna(thresh=5)
display("df2", "df5")df2
cold | slow | NaN | 2.0 | 6.0 | 3.0 |
|---|---|---|---|---|---|
warm | medium | 4.0 | 5.0 | 7.0 | 9.0 |
hot | fast | 9.0 | 4.0 | NaN | 6.0 |
cool | None | NaN | NaN | 17.0 | 89.0 |
cool | medium | 16.0 | 44.0 | 21.0 | 13.0 |
cold | slow | NaN | 29.0 | 33.0 | 17.0 |
df5
cold | slow | NaN | 2.0 | 6.0 | 3.0 |
|---|---|---|---|---|---|
warm | medium | 4.0 | 5.0 | 7.0 | 9.0 |
hot | fast | 9.0 | 4.0 | NaN | 6.0 |
cool | medium | 16.0 | 44.0 | 21.0 | 13.0 |
cold | slow | NaN | 29.0 | 33.0 | 17.0 |
Rather than dropping rows and columns, missing values can be imputed or replaced. The .fillna() method returns either a Series or a DataFrame with null values replaced. The example below replaces all NaN's with zero.
df6 = df2.fillna(0)
display("df2", "df6")df2
cold | slow | NaN | 2.0 | 6.0 | 3.0 |
|---|---|---|---|---|---|
warm | medium | 4.0 | 5.0 | 7.0 | 9.0 |
hot | fast | 9.0 | 4.0 | NaN | 6.0 |
cool | None | NaN | NaN | 17.0 | 89.0 |
cool | medium | 16.0 | 44.0 | 21.0 | 13.0 |
cold | slow | NaN | 29.0 | 33.0 | 17.0 |
df6
cold | slow | 0.0 | 2.0 | 6.0 | 3.0 |
|---|---|---|---|---|---|
warm | medium | 4.0 | 5.0 | 7.0 | 9.0 |
hot | fast | 9.0 | 4.0 | 0.0 | 6.0 |
cool | 0 | 0.0 | 0.0 | 17.0 | 89.0 |
cool | medium | 16.0 | 44.0 | 21.0 | 13.0 |
cold | slow | 0.0 | 29.0 | 33.0 | 17.0 |
As you can see from the example in the cell above, the .fillna() method is applied to all DataFrame cells. We may not wish to have missing values in df['col2'] replaced with zeros since they are strings. The method is applied to a list of target columns using the .loc method. The details for the .loc method are discussed in Chapter 05--Understanding Indexes .
df7 = df2[["col3", "col4", "col5", "col6"]].fillna(0)
display("df2", "df7")df2
cold | slow | NaN | 2.0 | 6.0 | 3.0 |
|---|---|---|---|---|---|
warm | medium | 4.0 | 5.0 | 7.0 | 9.0 |
hot | fast | 9.0 | 4.0 | NaN | 6.0 |
cool | None | NaN | NaN | 17.0 | 89.0 |
cool | medium | 16.0 | 44.0 | 21.0 | 13.0 |
cold | slow | NaN | 29.0 | 33.0 | 17.0 |
df7
0.0 | 2.0 | 6.0 | 3.0 |
|---|---|---|---|
4.0 | 5.0 | 7.0 | 9.0 |
9.0 | 4.0 | 0.0 | 6.0 |
0.0 | 0.0 | 17.0 | 89.0 |
16.0 | 44.0 | 21.0 | 13.0 |
0.0 | 29.0 | 33.0 | 17.0 |
An imputation method based on the mean value of df['col6'] is shown below. The .fillna() method finds and then replaces all occurrences of NaN with this calculated value.
df8 = df2[["col3", "col4", "col5"]].fillna(df2.col6.mean())
display("df2", "df8")df2
cold | slow | NaN | 2.0 | 6.0 | 3.0 |
|---|---|---|---|---|---|
warm | medium | 4.0 | 5.0 | 7.0 | 9.0 |
hot | fast | 9.0 | 4.0 | NaN | 6.0 |
cool | None | NaN | NaN | 17.0 | 89.0 |
cool | medium | 16.0 | 44.0 | 21.0 | 13.0 |
cold | slow | NaN | 29.0 | 33.0 | 17.0 |
df8
22.833333 | 2.000000 | 6.000000 |
|---|---|---|
4.000000 | 5.000000 | 7.000000 |
9.000000 | 4.000000 | 22.833333 |
22.833333 | 22.833333 | 17.000000 |
16.000000 | 44.000000 | 21.000000 |
22.833333 | 29.000000 | 33.000000 |
The corresponding SAS program is shown below. The PROC SQL SELECT INTO clause stores the calculated mean for the variable col6 into the macro variable &col6_mean. This is followed by a Data Step iterating the array x for col3 - col5 and replacing missing values with &col6_mean.
SAS/Stat has PROC MI for imputation of missing values with a range of methods described
here. PROC MI is outside the scope of these examples.
The .fillna(method='ffill') is a 'forward' fill method. NaN's are replaced by the adjacent cell above traversing 'down' the columns. The cell below contrasts the DataFrame df2, created above with the DataFrame df9 created with the 'forward' fill method.
df9 = df2.fillna(method='ffill')
display("df2", "df9")df2
cold | slow | NaN | 2.0 | 6.0 | 3.0 |
|---|---|---|---|---|---|
warm | medium | 4.0 | 5.0 | 7.0 | 9.0 |
hot | fast | 9.0 | 4.0 | NaN | 6.0 |
cool | None | NaN | NaN | 17.0 | 89.0 |
cool | medium | 16.0 | 44.0 | 21.0 | 13.0 |
cold | slow | NaN | 29.0 | 33.0 | 17.0 |
df9
cold | slow | NaN | 2.0 | 6.0 | 3.0 |
|---|---|---|---|---|---|
warm | medium | 4.0 | 5.0 | 7.0 | 9.0 |
hot | fast | 9.0 | 4.0 | 7.0 | 6.0 |
cool | fast | 9.0 | 4.0 | 17.0 | 89.0 |
cool | medium | 16.0 | 44.0 | 21.0 | 13.0 |
cold | slow | 16.0 | 29.0 | 33.0 | 17.0 |
Similarly, the.fillna(bfill) is a 'backwards' fill method. NaN's are replaced by the adjacent cell traversing 'up' the columns. The cell below contrasts the DataFrame df2, created above with the DataFrame df10 created with the 'backward' fill method.
pythondf10 = df2.fillna(method="bfill")display("df2", "df10")df2
cold | slow | NaN | 2.0 | 6.0 | 3.0 |
|---|---|---|---|---|---|
warm | medium | 4.0 | 5.0 | 7.0 | 9.0 |
hot | fast | 9.0 | 4.0 | NaN | 6.0 |
cool | None | NaN | NaN | 17.0 | 89.0 |
cool | medium | 16.0 | 44.0 | 21.0 | 13.0 |
cold | slow | NaN | 29.0 | 33.0 | 17.0 |
df10
cold | slow | 4.0 | 2.0 | 6.0 | 3.0 |
|---|---|---|---|---|---|
warm | medium | 4.0 | 5.0 | 7.0 | 9.0 |
hot | fast | 9.0 | 4.0 | 17.0 | 6.0 |
cool | medium | 16.0 | 44.0 | 17.0 | 89.0 |
cool | medium | 16.0 | 44.0 | 21.0 | 13.0 |
cold | slow | NaN | 29.0 | 33.0 | 17.0 |
Below we contrast DataFrame df9 created above using the 'forward' fill method, with DataFrame <code>df10</code> created with the 'backward' fill method.
display("df9", "df10")df9
cold | slow | NaN | 2.0 | 6.0 | 3.0 |
|---|---|---|---|---|---|
warm | medium | 4.0 | 5.0 | 7.0 | 9.0 |
hot | fast | 9.0 | 4.0 | 7.0 | 6.0 |
cool | fast | 9.0 | 4.0 | 17.0 | 89.0 |
cool | medium | 16.0 | 44.0 | 21.0 | 13.0 |
cold | slow | 16.0 | 29.0 | 33.0 | 17.0 |
df10
cold | slow | 4.0 | 2.0 | 6.0 | 3.0 |
|---|---|---|---|---|---|
warm | medium | 4.0 | 5.0 | 7.0 | 9.0 |
hot | fast | 9.0 | 4.0 | 17.0 | 6.0 |
cool | medium | 16.0 | 44.0 | 17.0 | 89.0 |
cool | medium | 16.0 | 44.0 | 21.0 | 13.0 |
cold | slow | NaN | 29.0 | 33.0 | 17.0 |
Before dropping the missing rows, calculate the portion of records lost in the accidents DataFrame, df created above.
print("{} records in the DataFrame will be dropped.".format(df.Time.isnull().sum()))
print("The portion of records dropped is {0:6.3%}".format(df.Time.isnull().sum() / (len(df) - df.Time.isnull().sum())))24 records in the DataFrame will be dropped.The portion of records dropped is 0.009%The .dropna() method is silent except in the case of errors. We can verify the DataFrame's shape after the method is applied.
print(df.shape)
df = df.dropna()
print(df.shape)(266776, 27)(266752, 27)Resources
10 Minutes to pandas from pandas.pydata.org.
Tutorials , and just below this link is the link for the pandas Cookbook, from the pandas 0.19.1 documentation at pandas.pydata.org.
pandas Home page for Python Data Analysis Library.
Python Data Science Handbook , Essential Tools for Working With Data, by Jake VanderPlas.
pandas: Data Handling and Analysis in Python from 2013 BYU MCL Bootcamp documentation.
Intro to pandas data structures by Greg Reda. This is a three-part series using the Movie Lens data set nicely to illustrate pandas.
Cheat Sheet: The pandas DataFrame Object by Mark Graph and located at the University of Idaho's web-site.
Working with missing data pandas 0.19.1 documentation.
Read the Book
This post is an excerpt from Randy Betancourt Python for SAS Users quick start guide. View the full Chapter List.
Randy Betancourt has spent his career in a number of customer and executive-facing roles at SAS Institute, Inc. and the Institute of International Analytics. Starting as a technical architect and more recently as a consultant, he advises business leaders on how to nurture and cost-effectively manage their analytical resources portfolio. Recently, these discussion and efforts have centered on modernization strategies in light of the growing industry innovations.



