The Importance of Structure, Coding Style, and Refactoring in Notebooks
Nikolay Manchev2020-07-01 | 26 min read

Notebooks are increasingly crucial in the data scientist's toolbox. Although considered relatively new, their history traces back to systems like Mathematica and MATLAB. This form of interactive workflow was introduced to assist data scientists in documenting their work, facilitating reproducibility, and prompting collaboration with their team members. Recently there has been an influx of newcomers, and data scientists now have a wide range of implementations to choose from, such as Jupyter Notebook, Zeppelin, R Markdown, Spark Notebook, and Polynote.
For Data Scientists, spinning up notebook instances as the first step in exploratory data analysis has become second nature. It's straight forward to get access to cloud compute, and the ability to mix code, outputs, and plots that notebooks offer is unparalleled. At a team level, notebooks can significantly enhance knowledge sharing, traceability, and accelerating the speed at which new insights can be discovered. To get the best out of notebooks, they must have good structure and follow good document and coding conventions.
In this article, I will talk about best practices to implement in your notebooks covering notebook structure, coding style, abstraction, and refactoring. The article concludes with an example of a notebook that implements these best practices.
Notebook Structure
Much like unstructured code, poorly organized notebooks can be hard to read and defeat the intended purpose of why you use notebooks, creating self-documenting readable code. Elements like markdown, concise code commentary and the use of section navigation help bring structure to notebooks, which enhance their potential for knowledge sharing and reproducibility.
Taking a step back and thinking what a good notebook looks like, we can look towards scientific papers for guidance. The hallmarks of an excellent scientific paper are clarity, simplicity, neutrality, accuracy, objectiveness, and above all - logical structure. Building a plan for your notebook before you start is a good idea. Opening an empty notebook and instantly typing your first import, i.e., "import numpy as np" is not the best way to go.
Here are a few simple steps that could encourage you to think about the big picture before your thoughts get absorbed by multidimensional arrays:
- Establish the purpose of your notebook. Think about the audience that you're aiming for and the specific problem you're trying to solve. What do your readers need to learn from it?
- In cases where there isn't a single straight forward goal, consider splitting work into multiple notebooks and creating a single master markdown document to explain the concept in its entirety, with links back to the related notebook.
- Use sections to help construct the right order of your notebook. Consider the best way to organize your work. For example, chronologically, where you start with exploration and data preparation before model training and evaluation. Alternatively, comparison/contrast sections that go into time, space, or sample complexity for comparing two different algorithms.
- Just like academic research papers, don't forget to include a title, preamble, table of contents, conclusion and reference any sources you've used in writing your code.
Adding a table of contents into your notebook is simple, using markdown and HTML. Here is a bare-bones example of a table of contents.
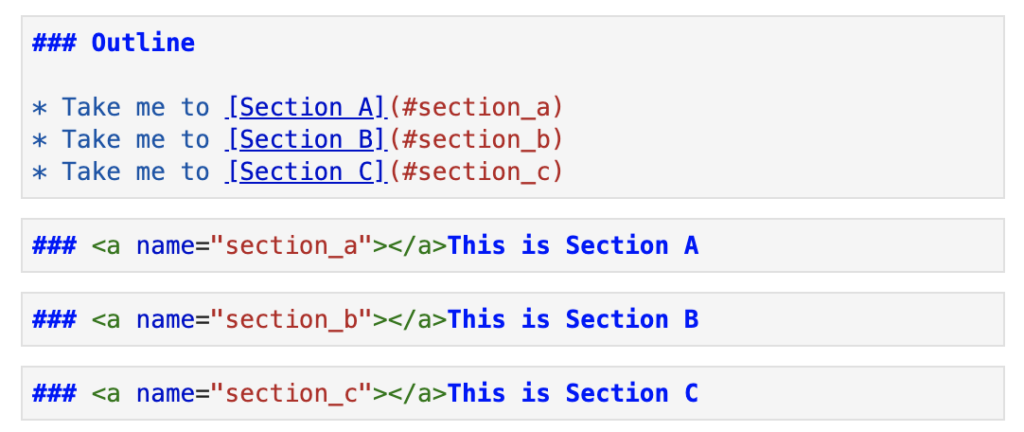
The rendered cells below give the notebook more structure and a more polished look. It makes the document easier to navigate and also helps to inform readers of the notebook structure at a single glance.

For further examples of good structure in notebooks, head to Kaggle and pick a random challenge, then look at the top five notebooks by user votes. These notebooks will be structured in a way that allows you to see the intended purpose and code flow without needing to run the code to understand it. If you compare the top five notebooks to any in the bottom ten percent, you'll see how important structure is in creating an easy to read notebook.
Code Style
"Code is read much more often than it is written" is a quote often attributed to the creator of Python, Guido van Rossum.
When you're collaborating in a team, adhering to an agreed style is essential. By following proper coding conventions, you create code that is more easily readable and therefore makes it easier for your colleagues to collaborate and help in any code review. Using consistent patterns to how variables are named and functions are called can also help you find obscure bugs. For example, using a lowercase L and an uppercase I in a variable name is a bad practice.
This section is written with Python in mind, but the principles outlined can also apply to other coding languages such as R. If you are interested in a useful convention guide for R, check R Style. An Rchaeological Commentary by Paul E. Johnson. A thorough discussion on what makes good code versus bad code is outside the scope of this article. The intention is to highlight some of the basics that often go unnoticed and highlight common offenders.
Readers are encouraged to familiarize themselves with Python Enhancement Proposal 8 (PEP-8) as it is referenced through this section and provides conventions on how to write good clear code.
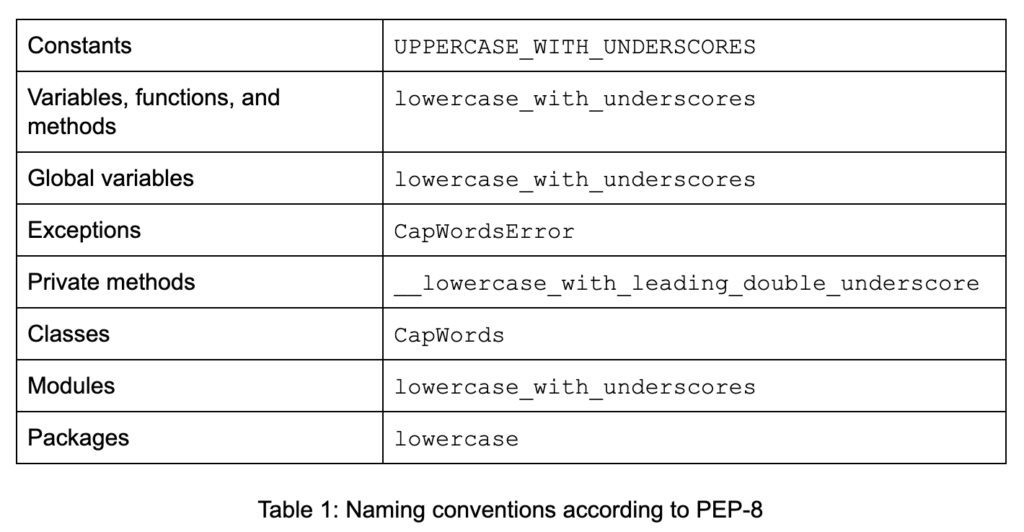
Constants, functions, variables, and other constructs in your code should have meaningful names and should adhere to naming conventions (see Table 1).
Naming things can be difficult. Often we are tempted to go for placeholder variable names such as "i" or "x" but consider that your code will be read far more times than it is written, and you'll understand the importance of proper names. Some good habits that I've picked up while coding in a dynamically-typed language like Python include
- Prefix variable types like boolean to make them more readable - i.e "is" or "has" - (
is_enabled,has_valueetc.) - Use plurals for arrays. For example,
book_namesversusbook_name - Describe numerical variables. Instead of just
ageconsideravg_ageormin_agedepending on the purpose of the variable.
Even if you don't go the extra mile and you don't follow PEP-8 to the letter, you should make an effort to adopt the majority of its recommendations. It is also very important to be consistent. Most people won't have an issue if you go for single or double quotes, and you can also get away with mixing them if you do it consistently (e.g. double quotes for strings, single quotes for regular expressions). But it can quickly get annoying if you have to read code that mixes them arbitrarily, and you will often catch yourself fixing quotes as you go along instead of focusing on the problem at hand.
Some things that are best avoided:
- Names with capitalised letters and underscores (e.g.
This_Is_Very_Ugly) - Non-ASCII characters and non-English identifiers in general. For example:
def 곱셈(일,이):
return 일*이- Abbreviations (e.g.
BfrWrtLmt)
When it comes to indentation tabs should be avoided - 4 spaces per indentation level is the recommended method. The PEP-8 guide also talks about the preferred way of aligning continuation lines. One violation that can be seen quite often is misalignment of arguments. For example:
foo = long_function_name(var_one, var_two
var_three, var_four)Instead of the much neater vertical alignment below.
foo = long_function_name(var_one, var_two
var_three, var_four)This is similar to line breaks, where we should aim to always break before binary operators.
# No: Operators sit far away from their operands
income = (gross_wages + taxable_interest + (dividends - qualified_dividends))
# Yes: Easy to match operators with operands
income = (gross_wages + taxable_interest + (dividends - qualified_dividends))Talking about line breaks logically leads us to a common offender that often sneaks its way into rushed code, namely blank lines. To be honest, I am also guilty of being too lenient around excessive use of blank lines. A good style demands that they should be used as follows:
- Surround top-level function and class definitions with two blank lines
- Method definitions inside a class are surrounded by a single blank line
- Use blank lines in functions (sparingly) to indicate logical sections
The key point in the last of the three is "sparingly". Leaving a blank or two around every other line in your code brings nothing to the table besides making you scroll up and down unnecessarily.
Last but not least, we need to say a few words about imports. There are only three important rules to watch for:
- Each import should be on a separate line (
import sys,osis a no-go) - The grouping order should be a standard library, third-party imports, and finally - local application
- Avoid wildcard imports (e.g.
from numpy import *), as they make it unclear which names are present in the namespace and often confuse automation tools
This may feel like a lot to take in, and after all, we're data scientists - not software engineers; but like any data scientist who has attempted to go back to work they did several years earlier, or pick up the pieces of a remaining project from another data scientist, implementing good structure and considering that others may read your code is important. Being able to share your work aids knowledge transfer and collaboration which in turn builds more success in data science practices. Having a shared style guide and code conventions in place promote effective teamwork and help set junior scientists up with an understanding of what success looks like.
Although your options for using code style checkers, or "code linters" as they are often called, is limited in Jupyter, there are certain extensions that could help. One extension that can be useful is pycodestyle. Its installation and usage is really straightforward. After installing and loading the extension you can put a %%pycodestyle magic command and pycodestyle outline any violations of PEP-8 that it can detect.
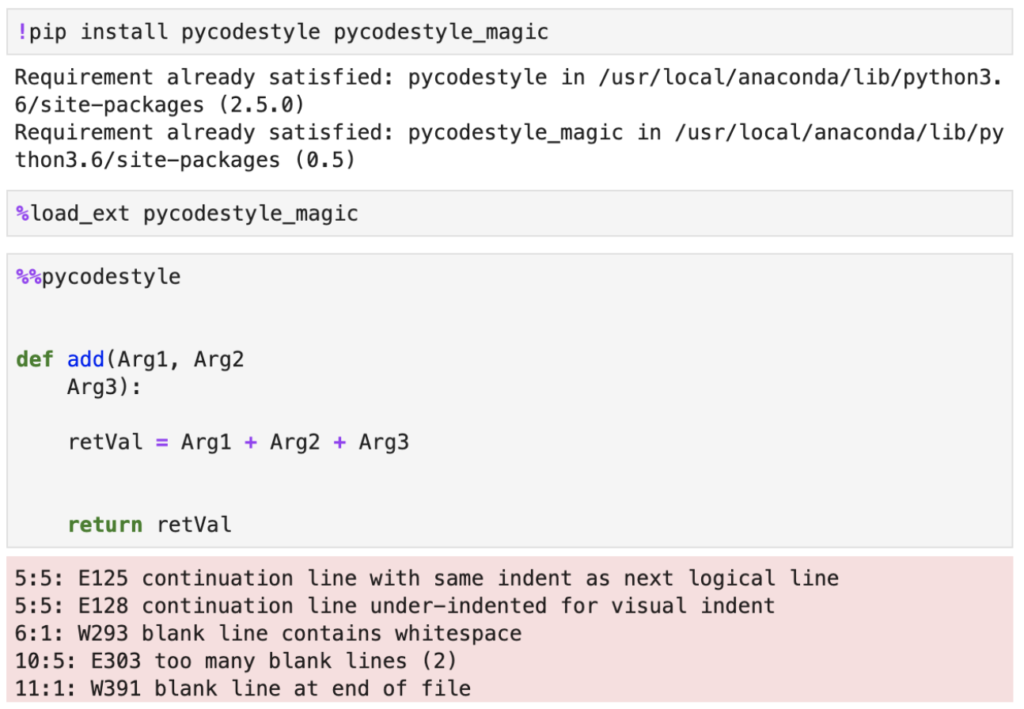
Note that you can also run pycodestyle from JupyterLab's terminal and analyze Python scripts, so the extension is not limited to notebooks only. It also has some neat features around showing you exact spots in your code where violations have been detected, plus it can compute statistics on different violation types.
Use of Abstractions and The Refactoring Process
Abstraction, or the idea of exposing only essential information and hiding complexity away, is a fundamental programming principle, and there is no reason why we shouldn't be applying it to Data Science-specific code.
Let's look at the following code snippet:
completeDF = dataDF[dataDF["poutcome"]!="unknown"]
completeDF = completeDF[completeDF["job"]!="unknown"]
completeDF = completeDF[completeDF["education"]!="unknown"]
completeDF = completeDF[completeDF["contact"]!="unknown"]The purpose of the code above is to remove any rows that contain the value "unknown" in any column of the DataFrame dataDF, and create a new DataFrame named completeDF that contains only complete cases.
This can easily be rewritten using an abstract function get_complete_cases() that's used in conjunction with dataDF. We might as well change the name of the resulting variable to conform to the PEP-8 style guide while we are at it.
def get_complete_cases(df):
return df[~df.eq("unknown").any(1)]
complete_df = get_complete_cases(dataDF)The benefits of using abstractions this way are easy to point out. Replacing repeated code with a function:
- Reduces the unnecessary duplication of code
- Promotes reusability (especially if we parametrize the value that defines a missing observation)
- Makes the code easier to test
- As a bonus point: Makes the code self-documenting. It's fairly straightforward to deduce what the content of
complete_dfwould be after seeing the function name and the argument that it receives
Many data scientists view notebooks as great for exploratory data analysis and communicating results with others but that they're not as useful when it comes to putting code into production; but increasingly notebooks are becoming a viable way to build production models. When looking to deploy a notebook that will become a scheduled production job, consider these options.
- Having an "orchestrator" notebook that uses the
%runmagic to execute other notebooks - Using tools like papermill, which allows parameterization and execution of notebooks from Python or via CLI. Scheduling can be fairly simple and maintained directly in crontab
- More complex pipelines that use Apache Airflow for orchestration and its papermill operator for the execution of notebooks. This option is quite powerful. It supports heterogeneous workflows and allows for conditional and parallel execution of notebooks
No matter what path to production we prefer for our code (extracting or directly orchestrating via notebook), the following general rules should be observed:
- Majority of the code should be in well-abstracted functions
- The functions should be placed in modules and packages
This brings us to notebook refactoring, which is a process every data scientist should be familiar with. No matter if we are aiming to get notebook code production-ready, or just want to push code out of the notebook and into modules, we can iteratively go over the following steps, which include both preparation and the actual refactoring:
- Restart the kernel and run all cells - it makes no sense to refactor a non-working notebook, so our first task is to make sure that the notebook doesn't depend on any hidden states and it cells can be successfully executed in sequence
- Make a copy of the notebook - it is easy to start refactoring and break the notebook to a point where you can't recover its original state. Working with a copy is a simpler alternative, and you also have the original notebook to get back to if something goes wrong
- Convert the notebook to Python code -
nbconvertprovides a simple and easy way of converting a notebook to an executable script. All you have to do is call it with your notebook's name:
$ jupyter nbconvert --to script- Tidy up the code - at this step, you might want to remove irrelevant cell outputs, transform cells to functions, remove markdown etc.
- Refactor - this is the main step in the process, where we restructure the existing body of code, altering its internal structure without changing its external behaviour. We will cover the refactoring cycle in details below
- [if needed] Repeat from step 5
[else] Restart the kernel and re-run all cells, making sure that the final notebook executes properly and performs as expected
Now let's dive into the details of Step 5.
The Refactor Step
The refactor step is a set of cyclic actions that can be repeated as many times as needed. The cycle starts with identifying a piece of code from the notebook that we want to extract. This code will be transformed to an external function and we need to write a unit test that comprehensively defines or improves the function in question. This approach is inspired by the test-driven development (TDD) process and also influenced by the test-first programming concepts of extreme programming.
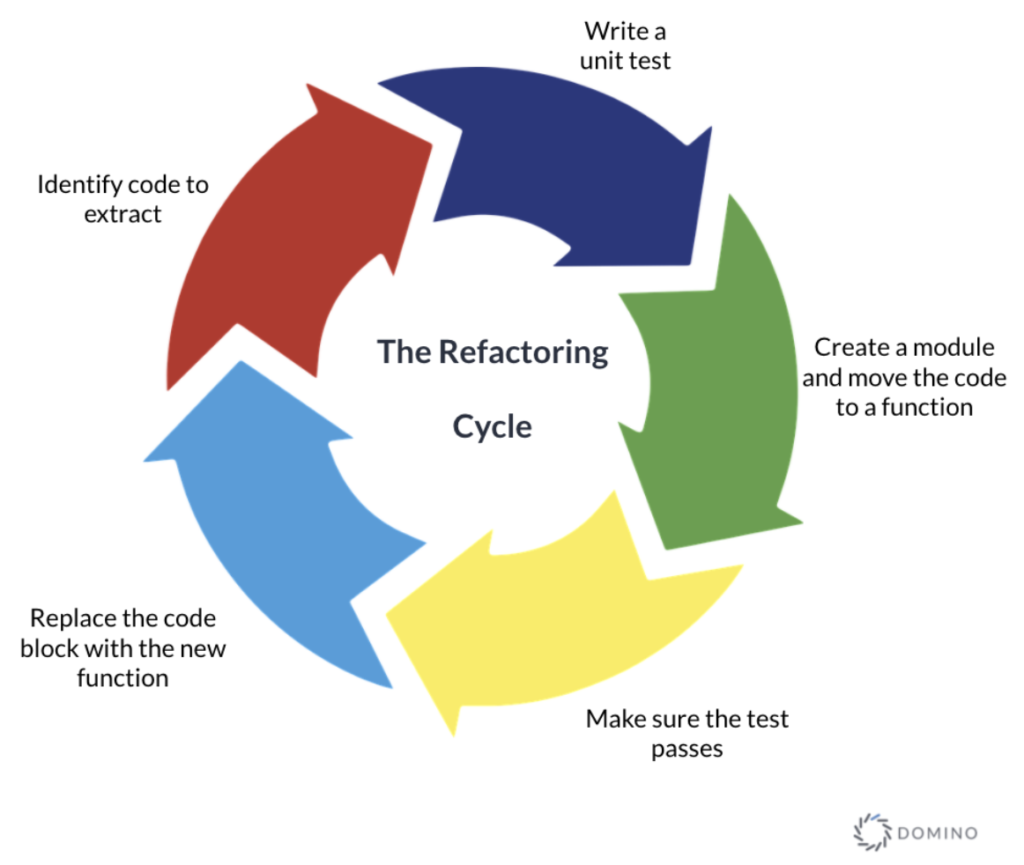
Figure 1 - The refactoring step cycle
Not every data scientist feels confident about code testing. This is especially true if they don't have a background in software engineering. Testing in Data Science, however, doesn't have to be at all complicated. It also brings tons of benefits by forcing us to be more mindful and results in code that is reliable, robust, and safe for reuse. When we think about testing in Data Science we usually consider two main types of tests:
- Unit tests that focus on individual units of source code. They are usually carried out by providing a simple input and observing the output of the code
- Integration tests that target integration of components. The objective here is to take a number of unit-tested components, combine them according to design specification, and test the output they produce
Sometimes an argument is brought up that due to its probabilistic nature machine learning code is not suitable for testing. This can't be further from the truth. First of all, plenty of workloads in a machine learning pipeline are fully deterministic (e.g. data processing). Second of all, we can always use metrics for non-deterministic workloads - think to measure the F1 score after fitting a binary classifier.
The most basic mechanism in Python is the assert statement. It is used to test a condition and immediately terminate the program if this condition is not met (see Figure 2).
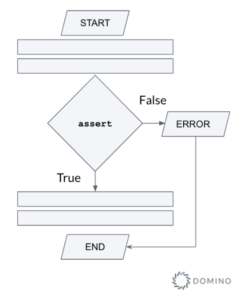
Figure 2 - Flowchart illustrating the use of the assert statement
The syntax of assert is
assert <statement>, <error>Generally, assert statements test for conditions that should never happen - that's why they immediately terminate the execution if the test statement evaluates to False. For example, to make sure that the function get_number_of_students() always returns non-negative values, you could add this to your code
assert get_number_of_students() &amp;gt;= 0, "Number of students cannot be negative."If for whatever reason the function returns a negative, your program will be terminated with a message similar to this:
Traceback (most recent call last):
File "", line xxx, in
AssertionError: Number of students cannot be negative.Asserts are helpful for basic checks in your code, and they can help you catch those pesky bugs, but they should not be experienced by users - that's what we have exceptions for. Remember, asserts are normally removed in release builds - they are not there to help the end-user, but they are key in assisting the developer and making sure that the code we produce is rock solid. If we are serious about the quality of our code, we should not only use asserts but adopt a comprehensive unit testing framework. The Python language includes unittest (often referred to as PyUnit) and this framework has been the de facto standard for testing Python code since Python 2.1. This is not the only testing framework available for Python (pytest and nose immediately spring to mind), but it is part of the standard library and there are some great tutorials to get you started. A test case unittest is created by subclassing unittest.TestCaseTests and tests are implemented as class methods. Each of the test cases calls one or more of the assertion methods provided by the framework (see Table 2).
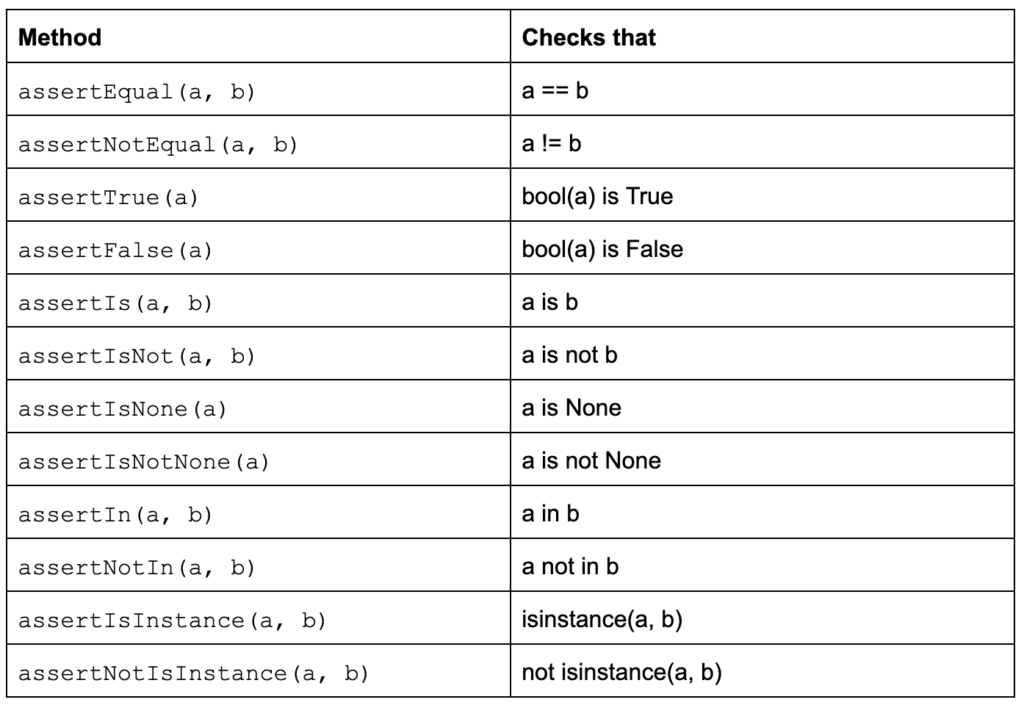
Table 2: TestCase class methods to check for and report failures
After we have a test class in place, we can proceed with creating a module and developing a Python function that passes the test case. We typically use the code from the nbconvert output but improve on it with the test case and reusability in mind. We then run the tests and confirm that our new function passes everything without issues. Finally, we replace the original code in the notebook copy with a call to the function and identify another piece of code to refactor.
We keep repeating the refactoring step as many times as needed until we end up with a tidy and concise notebook where the majority of the reusable code has been externalized. This entire process can be a lot to take in, so let's go over an end-to-end example that walks us over revamping a toy notebook.
An End-to-End Example
For this exercise we'll look at a very basic notebook (see Figure 3). Although this notebook is quite simplistic, it already has a table of contents (yay!), so someone has already thought about its structure or so we hope. The notebook, unoriginally named demo-notebook.ipynb, loads some CSV data into a Pandas DataFrame, displays the first 5 rows of data, and uses the snippet of code that we already looked at in the use of abstractions section to remove entries that contain the value "unknown" in four of the dataframe columns.
Following the process established above, we begin by restarting the kernel, followed by a Run All Cells command. After confirming that all cells execute correctly, we continue by creating a working copy of the notebook.
$ cp demo-notebook.ipynb demo-notebook-copy.ipynbNext, we convert the copy to a script using nbconvert.
$ jupyter nbconvert --to script demo-notebook-copy.ipynb
[NbConvertApp] Converting notebook demo-notebook.ipynb to script
[NbConvertApp] Writing 682 bytes to demo-notebook.py
$
Figure 3 - The sample notebook before refactoring
The result of nbconvert is a file named demo-notebook.py with the following contents:
#!/usr/bin/env python
# coding: utf-8
# ### Outline #
# * Take me to [Section A](#section_a)
# * Take me to [Section B](#section_b)
# * Take me to [Section C](#section_c)
# ### &amp;lt;a name="section_a"&amp;gt;&amp;lt;/a&amp;gt;This is Section A
import pandas as pd
dataDF = pd.read_csv("bank.csv")
dataDF.head()
# ### &amp;lt;a name="section_b"&amp;gt;&amp;lt;/a&amp;gt;This is Section B
completeDF = dataDF[dataDF["poutcome"]!="unknown"]
completeDF = completeDF[completeDF["job"]!="unknown"]
completeDF = completeDF[completeDF["education"]!="unknown"]
completeDF = completeDF[completeDF["contact"]!="unknown"]
completeDF.head()
# ### &amp;lt;a name="section_c"&amp;gt;&amp;lt;/a&amp;gt;This is Section CAt this point, we can rewrite the definition of completeDF as a function, and enclose the second head() call with a print call, so we can test the newly developed function. The Python script should now look like this (we omit the irrelevant parts for brevity).
...
def get_complete_cases(df):
return df[~df.eq("unknown").any(1)]
completeDF = get_complete_cases(dataDF)
print(completeDF.head())
...[/code]We can now run demo-notebook.py and confirm that the output is as expected.
$ python demo-notebook.py
age job marital education default balance housing ... month duration campaign pdays previous poutcome y
24060 33 admin. married tertiary no 882 no ... oct 39 1 151 3 failure no
24062 42 admin. single secondary no -247 yes ... oct 519 1 166 1 other yes
24064 33 services married secondary no 3444 yes ... oct 144 1 91 4 failure yes
24072 36 management married tertiary no 2415 yes ... oct 73 1 86 4 other no
24077 36 management married tertiary no 0 yes ... oct 140 1 143 3 failure yes[5 rows x 17 columns]Next, we proceed with the refactor step, targeting the get_complete_cases() function. Our first task after identifying the piece of code is to come up with a good set of tests that comprehensively test or improve the function. Here is a unit test that implements a couple of test cases for our function.
import unittest
import warnings
warnings.simplefilter(action="ignore", category=FutureWarning)
import numpy as np
import pandas as pd
from wrangler import get_complete_cases
class TestGetCompleteCases(unittest.TestCase):
def test_unknown_removal(self):
"""Test that it can sum a list of integers"""
c1 = [10, 1, 4, 5, 1, 9, 11, 15, 7, 83]
c2 = ["admin", "unknown", "services", "admin", "admin", "management", "unknown", "management", "services", "house-maid"]
c3 = ["tertiary", "unknown", "unknown", "tertiary", "secondary", "tertiary", "unknown", "unknown", "tertiary", "secondary"]
df = pd.DataFrame(list(zip(c1, c2, c3)), columns =["C1", "C2", "C3"])
complete_df = df[df["C2"]!="unknown"]
complete_df = complete_df[complete_df["C3"]!="unknown"]
complete_df_fn = get_complete_cases(df)
self.assertTrue(complete_df.equals(complete_df_fn))
def test_nan_removal(self):
"""Test that it can sum a list of integers"""
c1 = [10, 1, 4, 5, 1, np.nan, 11, 15, 7, 83]
c2 = ["admin", "services", "services", "admin", "admin", "management", np.nan, "management", "services", "house-maid"]
c3 = ["tertiary", "primary", "secondary", "tertiary", "secondary", "tertiary", np.nan, "primary", "tertiary", "secondary"]
df = pd.DataFrame(list(zip(c1, c2, c3)), columns =["C1", "C2", "C3"])
complete_df = df.dropna(axis = 0, how = "any")
complete_df_fn = get_complete_cases(df)
self.assertTrue(complete_df.equals(complete_df_fn))
if __name__ == '__main__':
unittest.main()
The code above reveals that I am planning to put get_complete_cases() in a package called wrangler. The second test case also makes it evident that I am planning to improve the in-scope function by also making it remove NaN's. You see that the way I carry out the tests is by constructing a DataFrame from a number of statically defined arrays. This is a rudimentary way of setting up tests and a better approach would be to leverage the setUp() and tearDown() methods of TestCase, so you might want to look at how to use those.
We can now move on to constructing our data wrangling module. This is done by creating a directory named wrangler and placing an __init.py__ file with the following contents in it:
import numpy as np
def get_complete_cases(df):
"""
Filters out incomplete cases from a Pandas DataFrame.
This function will go over a DataFrame and remove any row that contains the value "unknown"
or np.nan in any of its columns.
Parameters:
df (DataFrame): DataFrame to filter
Returns:
DataFrame: New DataFrame containing complete cases only
"""
return df.replace("unknown", np.nan).dropna(axis = 0, how = "any")You see from the code above that the function has been slightly modified to remove NaN entries as well. Time to see if the test cases pass successfully.
$ python test.py
..
----------------------------------------------------------------------
Ran 2 tests in 0.015sOKAfter getting a confirmation that all tests pass successfully, it is time to execute the final step and replace the notebook code with a call to the newly developed and tested function. The re-worked section of the notebook should look like this:
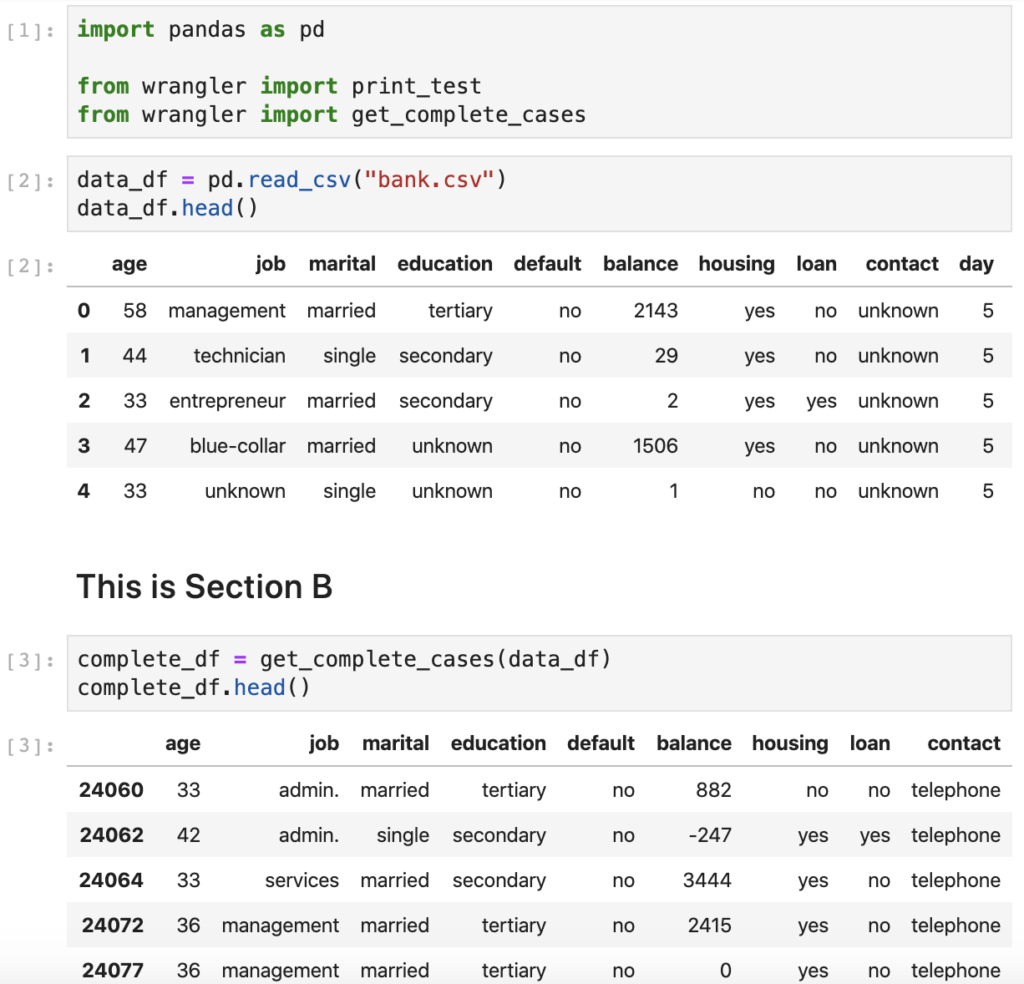
Since I am refactoring just the complete cases piece of code, I don't have to repeat the refactoring cycle any further. The final bit left to check is to bounce the kernel and make sure that all cells execute sequentially. As you can see above, the resulting notebook is concise, self-documenting, and generally looks better. In addition, we now have a standalone module that we can reuse across other notebooks and scripts.
Summary
Good software engineering practices can and should be applied to Data Science. There is nothing preventing us from developing notebook code that is readable, maintainable, and reliable. This article outlined some of the key principles that data scientists should adhere to when working on notebooks:
- Structure your content
- Observe a code style and be consistent
- Leverage abstractions
- Adopt a testing framework and develop a testing strategy for your code
- Refactor often and move the code to reusable modules
Machine Learning code and code generally written with data science applications in mind is no exception to the ninety-ninety rule. When writing code we should always consider its maintainability, dependability, efficiency, and usability. This article tried to outline some key principles for producing high-quality data science deliverables, but the elements covered are by no means exhaustive. Here is a brief list of additional habits and rules to be considered:
- Comments - not having comments is bad, but swinging the pendulum too far the other way doesn't help either. There is no value in comments that just repeat the code. Obvious code shouldn't be commented.
- DRY Principle (Don't Repeat Yourself) - repetitive code sections should be abstracted / automated.
- Deep nesting is evil - five nested ifs are hard to read and deep nesting is considered an anti-pattern.
- Project organization is key - Yes, you can do tons of things in a single notebook or a Python script, but having a logical directory structure and module organization helps tremendously, especially in complex projects.
- Version control is a must-have - if you often have to deal with notebooks whose names look like
notebook_5.ipynb,notebook_5_test_2.ipynb, ornotebook_2_final_v4.ipynb, you know something is not right. - Work in notebooks is notoriously hard to reproduce, as there are many elements that need to be considered (hardware, interpreter version, frameworks, and other libraries version, randomization control, source control, data integrity, etc.), but an effort should be made to at least store a
requirements.txtfile alongside each notebook.
Nikolay Manchev is a former Principal Data Scientist for EMEA at Domino Data Lab. In this role, Nikolay helped clients from a wide range of industries tackle challenging machine learning use-cases and successfully integrate predictive analytics in their domain-specific workflows. He holds an MSc in Software Technologies, an MSc in Data Science, and is currently undertaking postgraduate research at King's College London. His area of expertise is Machine Learning and Data Science, and his research interests are in neural networks and computational neurobiology.



